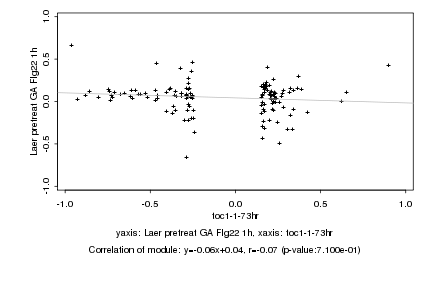
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
toc1-1-73hr |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.894 |
0.433 |
| 2 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.648 |
0.116 |
| 3 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.622 |
0.006 |
| 4 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
0.423 |
-0.121 |
| 5 |
249128_at |
AT5G43440
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.383 |
0.146 |
| 6 |
256593_at |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.369 |
0.304 |
| 7 |
262452_at |
AT1G11210
|
unknown |
0.362 |
0.155 |
| 8 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
0.340 |
-0.090 |
| 9 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.337 |
0.132 |
| 10 |
250381_at |
AT5G11610
|
[Exostosin family protein] |
0.333 |
-0.325 |
| 11 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.321 |
0.161 |
| 12 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
0.318 |
-0.162 |
| 13 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.314 |
0.115 |
| 14 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
0.303 |
-0.326 |
| 15 |
246095_at |
AT5G19310
|
CHR23, chromatin remodeling 23 |
0.281 |
-0.061 |
| 16 |
263128_at |
AT1G78600
|
BBX22, B-box domain protein 22, DBB3, DOUBLE B-BOX 3, LZF1, light-regulated zinc finger protein 1, STH3, SALT TOLERANCE HOMOLOG 3 |
0.281 |
0.135 |
| 17 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.272 |
0.103 |
| 18 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
0.269 |
0.060 |
| 19 |
255980_at |
AT1G33970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.256 |
-0.009 |
| 20 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
0.256 |
-0.488 |
| 21 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.243 |
-0.242 |
| 22 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
0.240 |
0.043 |
| 23 |
265713_at |
AT2G03530
|
ATUPS2, ARABIDOPSIS THALIANA UREIDE PERMEASE 2, UPS2, ureide permease 2 |
0.234 |
-0.011 |
| 24 |
246568_at |
AT5G14960
|
DEL2, DP-E2F-like 2, E2FD, E2L1 |
0.232 |
0.097 |
| 25 |
247532_at |
AT5G61560
|
[U-box domain-containing protein kinase family protein] |
0.231 |
0.045 |
| 26 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.229 |
0.044 |
| 27 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.228 |
0.108 |
| 28 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.228 |
0.071 |
| 29 |
260425_at |
AT1G72440
|
EDA25, embryo sac development arrest 25, SWA2, SLOW WALKER2 |
0.223 |
-0.095 |
| 30 |
254894_at |
AT4G11840
|
PLDGAMMA3, phospholipase D gamma 3 |
0.222 |
0.083 |
| 31 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.219 |
0.263 |
| 32 |
263182_at |
AT1G05575
|
unknown |
0.218 |
0.010 |
| 33 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
0.217 |
-0.092 |
| 34 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.215 |
-0.016 |
| 35 |
248321_at |
AT5G52740
|
[Copper transport protein family] |
0.211 |
0.124 |
| 36 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
0.208 |
0.072 |
| 37 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
0.206 |
0.029 |
| 38 |
265918_at |
AT2G15090
|
KCS8, 3-ketoacyl-CoA synthase 8 |
0.205 |
0.046 |
| 39 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.203 |
0.112 |
| 40 |
256170_at |
AT1G51790
|
[Leucine-rich repeat protein kinase family protein] |
0.203 |
0.078 |
| 41 |
249422_at |
AT5G39760
|
AtHB23, homeobox protein 23, HB23, homeobox protein 23, ZHD10, ZINC FINGER HOMEODOMAIN 10 |
0.200 |
0.093 |
| 42 |
267268_at |
AT2G02570
|
[nucleic acid binding] |
0.200 |
0.192 |
| 43 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
0.198 |
-0.216 |
| 44 |
259534_at |
AT1G12290
|
[Disease resistance protein (CC-NBS-LRR class) family] |
0.187 |
0.413 |
| 45 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.184 |
0.136 |
| 46 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
0.181 |
0.178 |
| 47 |
245600_at |
AT4G14230
|
unknown |
0.179 |
0.233 |
| 48 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
0.173 |
0.203 |
| 49 |
257191_at |
AT3G13175
|
unknown |
0.173 |
0.154 |
| 50 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
0.169 |
0.152 |
| 51 |
262913_at |
AT1G59960
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.169 |
-0.309 |
| 52 |
254043_at |
AT4G25990
|
CIL |
0.169 |
-0.113 |
| 53 |
266140_at |
AT2G28120
|
[Major facilitator superfamily protein] |
0.167 |
-0.028 |
| 54 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.167 |
0.180 |
| 55 |
260975_at |
AT1G53430
|
[Leucine-rich repeat transmembrane protein kinase] |
0.167 |
0.114 |
| 56 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
0.163 |
0.185 |
| 57 |
247216_at |
AT5G64860
|
DPE1, disproportionating enzyme |
0.162 |
-0.084 |
| 58 |
260671_at |
AT1G19310
|
[RING/U-box superfamily protein] |
0.161 |
0.170 |
| 59 |
247444_at |
AT5G62630
|
HIPL2, hipl2 protein precursor |
0.161 |
0.202 |
| 60 |
262784_at |
AT1G10760
|
GWD, GWD1, SEX1, STARCH EXCESS 1, SOP, SOP1 |
0.159 |
-0.228 |
| 61 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
0.156 |
-0.004 |
| 62 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
0.156 |
-0.436 |
| 63 |
257473_at |
AT1G33840
|
unknown |
0.155 |
0.091 |
| 64 |
260987_at |
AT1G53590
|
NTMC2T6.1, NTMC2TYPE6.1 |
0.155 |
0.073 |
| 65 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
0.154 |
-0.291 |
| 66 |
263118_at |
AT1G03090
|
MCCA |
0.153 |
0.051 |
| 67 |
255599_at |
AT4G01010
|
ATCNGC13, CYCLIC NUCLEOTIDE-GATED CHANNEL 13, CNGC13, cyclic nucleotide-gated channel 13 |
0.152 |
0.178 |
| 68 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.152 |
-0.039 |
| 69 |
250604_at |
AT5G07830
|
AtGUS2, glucuronidase 2, GUS2, glucuronidase 2 |
0.151 |
-0.140 |
| 70 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-0.967 |
0.668 |
| 71 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.933 |
0.029 |
| 72 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.886 |
0.079 |
| 73 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.861 |
0.120 |
| 74 |
250778_at |
AT5G05500
|
MOP10 |
-0.805 |
0.052 |
| 75 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.748 |
0.150 |
| 76 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.744 |
0.119 |
| 77 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.738 |
0.019 |
| 78 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.733 |
0.077 |
| 79 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-0.727 |
0.056 |
| 80 |
258751_at |
AT3G05890
|
RCI2B, RARE-COLD-INDUCIBLE 2B |
-0.714 |
0.118 |
| 81 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
-0.677 |
0.088 |
| 82 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.653 |
0.098 |
| 83 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.621 |
0.071 |
| 84 |
252833_at |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
-0.612 |
0.133 |
| 85 |
259276_at |
AT3G01190
|
[Peroxidase superfamily protein] |
-0.610 |
0.038 |
| 86 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-0.591 |
0.135 |
| 87 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-0.573 |
0.083 |
| 88 |
265050_at |
AT1G52070
|
[Mannose-binding lectin superfamily protein] |
-0.561 |
0.087 |
| 89 |
247337_at |
AT5G63660
|
LCR74, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 74, PDF2.5 |
-0.533 |
0.095 |
| 90 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
-0.519 |
0.051 |
| 91 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
-0.471 |
0.136 |
| 92 |
249375_at |
AT5G40730
|
AGP24, arabinogalactan protein 24, ATAGP24, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEIN 24 |
-0.471 |
0.022 |
| 93 |
253582_at |
AT4G30670
|
[Putative membrane lipoprotein] |
-0.466 |
0.456 |
| 94 |
261099_at |
AT1G62980
|
ATEXP18, ATEXPA18, expansin A18, ATHEXP ALPHA 1.25, EXP18, EXPANSIN 18, EXPA18, expansin A18 |
-0.462 |
0.039 |
| 95 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
-0.462 |
0.078 |
| 96 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.410 |
0.115 |
| 97 |
247965_at |
AT5G56540
|
AGP14, arabinogalactan protein 14, ATAGP14 |
-0.407 |
-0.112 |
| 98 |
264577_at |
AT1G05260
|
RCI3, RARE COLD INDUCIBLE GENE 3, RCI3A |
-0.391 |
0.146 |
| 99 |
251843_x_at |
AT3G54590
|
ATHRGP1, hydroxyproline-rich glycoprotein, HRGP1, hydroxyproline-rich glycoprotein |
-0.386 |
0.155 |
| 100 |
250696_at |
AT5G06790
|
unknown |
-0.374 |
-0.131 |
| 101 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.367 |
-0.050 |
| 102 |
245967_at |
AT5G19800
|
[hydroxyproline-rich glycoprotein family protein] |
-0.361 |
0.078 |
| 103 |
257924_at |
AT3G23190
|
[HR-like lesion-inducing protein-related] |
-0.357 |
0.124 |
| 104 |
266591_at |
AT2G46225
|
ABIL1, ABI-1-like 1 |
-0.354 |
-0.097 |
| 105 |
258765_at |
AT3G10710
|
RHS12, root hair specific 12 |
-0.348 |
0.062 |
| 106 |
251918_at |
AT3G54040
|
[PAR1 protein] |
-0.326 |
0.400 |
| 107 |
259964_at |
AT1G53680
|
ATGSTU28, glutathione S-transferase TAU 28, GSTU28, glutathione S-transferase TAU 28 |
-0.322 |
0.098 |
| 108 |
250469_at |
AT5G10130
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.322 |
0.100 |
| 109 |
257481_at |
AT1G08430
|
ALMT1, aluminum-activated malate transporter 1, ATALMT1, ARABIDOPSIS THALIANA ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 |
-0.319 |
0.066 |
| 110 |
246125_at |
AT5G19875
|
unknown |
-0.302 |
-0.219 |
| 111 |
258080_at |
AT3G25930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.292 |
0.092 |
| 112 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.292 |
-0.654 |
| 113 |
248652_at |
AT5G49270
|
COBL9, COBRA-LIKE 9, DER9, DEFORMED ROOT HAIRS 9, MRH4, MUTANT ROOT HAIR 4, SHV2, SHAVEN 2 |
-0.291 |
0.077 |
| 114 |
264029_at |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
-0.291 |
0.041 |
| 115 |
250323_at |
AT5G12880
|
[proline-rich family protein] |
-0.289 |
0.160 |
| 116 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
-0.288 |
0.079 |
| 117 |
247024_at |
AT5G66985
|
unknown |
-0.286 |
-0.095 |
| 118 |
254107_at |
AT4G25220
|
AtG3Pp2, glycerol-3-phosphate permease 2, G3Pp2, glycerol-3-phosphate permease 2, RHS15, root hair specific 15 |
-0.285 |
0.066 |
| 119 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
-0.278 |
0.214 |
| 120 |
250632_at |
AT5G07450
|
CYCP4;3, cyclin p4;3 |
-0.278 |
0.153 |
| 121 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
-0.276 |
-0.213 |
| 122 |
256674_at |
AT3G52360
|
unknown |
-0.276 |
-0.024 |
| 123 |
245113_at |
AT2G41660
|
MIZ1, mizu-kussei 1 |
-0.276 |
0.281 |
| 124 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.272 |
0.154 |
| 125 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
-0.271 |
-0.051 |
| 126 |
246519_at |
AT5G15780
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.266 |
0.102 |
| 127 |
260030_at |
AT1G68880
|
AtbZIP, basic leucine-zipper 8, bZIP, basic leucine-zipper 8 |
-0.265 |
0.052 |
| 128 |
261930_at |
AT1G22440
|
[Zinc-binding alcohol dehydrogenase family protein] |
-0.261 |
0.053 |
| 129 |
266581_at |
AT2G46140
|
[Late embryogenesis abundant protein] |
-0.260 |
0.358 |
| 130 |
258184_at |
AT3G21510
|
AHP1, histidine-containing phosphotransmitter 1 |
-0.259 |
-0.190 |
| 131 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.258 |
0.068 |
| 132 |
259478_at |
AT1G18980
|
[RmlC-like cupins superfamily protein] |
-0.256 |
0.466 |
| 133 |
247678_at |
AT5G59520
|
AtZIP2, ZIP2, ZRT/IRT-like protein 2 |
-0.254 |
0.078 |
| 134 |
263614_at |
AT2G25240
|
AtCCP3, CCP3, conserved in ciliated species and in the land plants 3 |
-0.254 |
0.039 |
| 135 |
256192_at |
AT1G30110
|
ATNUDX25, nudix hydrolase homolog 25, NUDX25, nudix hydrolase homolog 25 |
-0.252 |
-0.195 |
| 136 |
250002_at |
AT5G18690
|
AGP25, arabinogalactan protein 25, ATAGP25, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEINS 25 |
-0.249 |
-0.103 |
| 137 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.246 |
-0.362 |