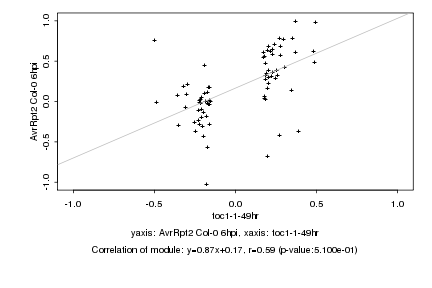
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
toc1-1-49hr |
Log2 signal ratio
AvrRpt2 Col-0 6hpi |
| 1 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
0.493 |
0.989 |
| 2 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.483 |
0.484 |
| 3 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.478 |
0.628 |
| 4 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.388 |
-0.365 |
| 5 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.369 |
0.614 |
| 6 |
261249_at |
AT1G05880
|
ARI12, ARIADNE 12, ATARI12, ARABIDOPSIS ARIADNE 12 |
0.369 |
1.003 |
| 7 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.348 |
0.787 |
| 8 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.342 |
0.144 |
| 9 |
246978_at |
AT5G24910
|
CYP714A1, cytochrome P450, family 714, subfamily A, polypeptide 1, ELA1, EUI-like p450 A1 |
0.299 |
0.426 |
| 10 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.294 |
0.776 |
| 11 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.276 |
0.688 |
| 12 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.275 |
0.573 |
| 13 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
0.268 |
-0.416 |
| 14 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.267 |
0.789 |
| 15 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.254 |
0.325 |
| 16 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.250 |
0.391 |
| 17 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.241 |
0.296 |
| 18 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.235 |
0.708 |
| 19 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
0.228 |
0.649 |
| 20 |
255923_at |
AT1G22180
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.225 |
0.361 |
| 21 |
259921_at |
AT1G72540
|
[Protein kinase superfamily protein] |
0.223 |
0.594 |
| 22 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.219 |
0.314 |
| 23 |
265597_at |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.216 |
0.621 |
| 24 |
260744_at |
AT1G15010
|
unknown |
0.201 |
0.685 |
| 25 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
0.199 |
0.227 |
| 26 |
260425_at |
AT1G72440
|
EDA25, embryo sac development arrest 25, SWA2, SLOW WALKER2 |
0.198 |
0.307 |
| 27 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.198 |
0.391 |
| 28 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
0.197 |
-0.677 |
| 29 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.196 |
0.640 |
| 30 |
254248_at |
AT4G23270
|
CRK19, cysteine-rich RLK (RECEPTOR-like protein kinase) 19 |
0.194 |
0.172 |
| 31 |
260996_at |
AT1G12210
|
RFL1, RPS5-like 1 |
0.189 |
0.353 |
| 32 |
261973_at |
AT1G64610
|
[Transducin/WD40 repeat-like superfamily protein] |
0.184 |
0.472 |
| 33 |
260394_at |
AT1G74080
|
ATMYB122, MYB DOMAIN PROTEIN 122, MYB122, myb domain protein 122 |
0.184 |
0.329 |
| 34 |
263128_at |
AT1G78600
|
BBX22, B-box domain protein 22, DBB3, DOUBLE B-BOX 3, LZF1, light-regulated zinc finger protein 1, STH3, SALT TOLERANCE HOMOLOG 3 |
0.182 |
0.032 |
| 35 |
259911_at |
AT1G72680
|
ATCAD1, CINNAMYL ALCOHOL DEHYDROGENASE 1, CAD1, cinnamyl-alcohol dehydrogenase |
0.180 |
0.282 |
| 36 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
0.178 |
0.066 |
| 37 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.177 |
0.563 |
| 38 |
259320_at |
AT3G01080
|
ATWRKY58, WRKY DNA-BINDING PROTEIN 58, WRKY58, WRKY DNA-binding protein 58 |
0.176 |
0.046 |
| 39 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.173 |
0.615 |
| 40 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
0.173 |
0.551 |
| 41 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.504 |
0.758 |
| 42 |
248252_at |
AT5G53250
|
AGP22, arabinogalactan protein 22, ATAGP22, ARABINOGALACTAN PROTEIN 22 |
-0.492 |
-0.002 |
| 43 |
253957_at |
AT4G26320
|
AGP13, arabinogalactan protein 13 |
-0.361 |
0.085 |
| 44 |
266591_at |
AT2G46225
|
ABIL1, ABI-1-like 1 |
-0.354 |
-0.286 |
| 45 |
246125_at |
AT5G19875
|
unknown |
-0.324 |
0.187 |
| 46 |
253243_at |
AT4G34560
|
unknown |
-0.312 |
-0.066 |
| 47 |
264029_at |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
-0.308 |
0.099 |
| 48 |
261999_at |
AT1G33800
|
AtGXMT1, GXM1, glucuronoxylan methyltransferase 1, GXMT1, glucuronoxylan methyltransferase 1 |
-0.297 |
0.215 |
| 49 |
263513_at |
AT2G12400
|
unknown |
-0.255 |
-0.248 |
| 50 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.248 |
-0.371 |
| 51 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.234 |
-0.228 |
| 52 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.228 |
-0.103 |
| 53 |
257824_at |
AT3G25290
|
[Auxin-responsive family protein] |
-0.226 |
0.018 |
| 54 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.224 |
-0.279 |
| 55 |
256674_at |
AT3G52360
|
unknown |
-0.223 |
-0.004 |
| 56 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
-0.218 |
0.036 |
| 57 |
249009_at |
AT5G44610
|
MAP18, microtubule-associated protein 18, PCAP2, PLASMA MEMBRANE ASSOCIATED CA2+-BINDING PROTEIN-2 |
-0.217 |
-0.011 |
| 58 |
257081_at |
AT3G30460
|
[RING/U-box superfamily protein] |
-0.216 |
-0.024 |
| 59 |
256875_at |
AT3G26330
|
CYP71B37, cytochrome P450, family 71, subfamily B, polypeptide 37 |
-0.212 |
-0.093 |
| 60 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.211 |
-0.196 |
| 61 |
257026_at |
AT3G19200
|
unknown |
-0.210 |
0.062 |
| 62 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.207 |
-0.298 |
| 63 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-0.199 |
-0.432 |
| 64 |
260983_at |
AT1G53560
|
[Ribosomal protein L18ae family] |
-0.198 |
-0.135 |
| 65 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
-0.197 |
0.455 |
| 66 |
258143_at |
AT3G18170
|
[Glycosyltransferase family 61 protein] |
-0.193 |
0.105 |
| 67 |
263775_at |
AT2G46410
|
CPC, CAPRICE |
-0.189 |
0.003 |
| 68 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.182 |
-0.182 |
| 69 |
265892_at |
AT2G15020
|
unknown |
-0.180 |
-1.016 |
| 70 |
253096_at |
AT4G37330
|
CYP81D4, cytochrome P450, family 81, subfamily D, polypeptide 4 |
-0.178 |
-0.559 |
| 71 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
-0.177 |
-0.021 |
| 72 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.173 |
0.124 |
| 73 |
260366_at |
AT1G70460
|
AtPERK13, proline-rich extensin-like receptor kinase 13, PERK13, proline-rich extensin-like receptor kinase 13, RHS10, root hair specific 10 |
-0.170 |
-0.027 |
| 74 |
264894_at |
AT1G23040
|
[hydroxyproline-rich glycoprotein family protein] |
-0.169 |
0.182 |
| 75 |
254085_at |
AT4G24960
|
ATHVA22D, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE D, HVA22D, HVA22 homologue D |
-0.166 |
0.176 |
| 76 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.166 |
0.020 |
| 77 |
246063_at |
AT5G19340
|
unknown |
-0.164 |
-0.026 |
| 78 |
256603_at |
AT3G28270
|
unknown |
-0.161 |
-0.282 |
| 79 |
255587_at |
AT4G01480
|
AtPPa5, pyrophosphorylase 5, PPa5, pyrophosphorylase 5 |
-0.154 |
0.008 |