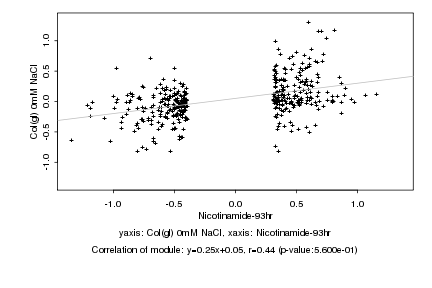
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Nicotinamide-93hr |
Log2 signal ratio
Col(gl) 0mM NaCl |
| 1 |
252511_at |
AT3G46280
|
[protein kinase-related] |
1.148 |
0.126 |
| 2 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
1.062 |
0.105 |
| 3 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
0.974 |
-0.003 |
| 4 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.944 |
0.041 |
| 5 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.895 |
0.228 |
| 6 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.888 |
0.110 |
| 7 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.867 |
-0.184 |
| 8 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.864 |
0.303 |
| 9 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.864 |
-0.006 |
| 10 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.845 |
0.405 |
| 11 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.832 |
0.087 |
| 12 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.809 |
1.179 |
| 13 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.803 |
0.009 |
| 14 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.794 |
0.001 |
| 15 |
248563_at |
AT5G49690
|
[UDP-Glycosyltransferase superfamily protein] |
0.793 |
-0.006 |
| 16 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.791 |
0.091 |
| 17 |
264375_at |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
0.789 |
0.019 |
| 18 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.750 |
0.151 |
| 19 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.746 |
0.020 |
| 20 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.740 |
1.040 |
| 21 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.716 |
-0.070 |
| 22 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.714 |
0.778 |
| 23 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.711 |
0.664 |
| 24 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
0.698 |
1.167 |
| 25 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.684 |
0.012 |
| 26 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
0.680 |
-0.129 |
| 27 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.679 |
0.406 |
| 28 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.676 |
1.153 |
| 29 |
254410_at |
AT4G21410
|
CRK29, cysteine-rich RLK (RECEPTOR-like protein kinase) 29 |
0.675 |
0.155 |
| 30 |
263475_at |
AT2G31945
|
unknown |
0.671 |
0.321 |
| 31 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.671 |
0.458 |
| 32 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.669 |
0.647 |
| 33 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
0.666 |
0.083 |
| 34 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.662 |
-0.056 |
| 35 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
0.655 |
0.673 |
| 36 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.654 |
-0.391 |
| 37 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
0.651 |
-0.000 |
| 38 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
0.625 |
0.249 |
| 39 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
0.621 |
0.054 |
| 40 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.620 |
-0.037 |
| 41 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
0.619 |
0.058 |
| 42 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.619 |
0.861 |
| 43 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
0.616 |
0.344 |
| 44 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
0.614 |
0.155 |
| 45 |
256170_at |
AT1G51790
|
[Leucine-rich repeat protein kinase family protein] |
0.613 |
0.274 |
| 46 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.609 |
0.075 |
| 47 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
0.604 |
0.582 |
| 48 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.602 |
-0.494 |
| 49 |
260926_at |
AT1G21360
|
GLTP2, glycolipid transfer protein 2 |
0.601 |
0.032 |
| 50 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.601 |
0.714 |
| 51 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.601 |
0.614 |
| 52 |
259511_at |
AT1G12520
|
ATCCS, copper chaperone for SOD1, CCS, copper chaperone for SOD1 |
0.597 |
0.212 |
| 53 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.591 |
1.314 |
| 54 |
256098_at |
AT1G13700
|
PGL1, 6-phosphogluconolactonase 1 |
0.590 |
0.569 |
| 55 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.584 |
0.264 |
| 56 |
263194_at |
AT1G36060
|
[Integrase-type DNA-binding superfamily protein] |
0.583 |
0.029 |
| 57 |
246418_at |
AT5G16960
|
[Zinc-binding dehydrogenase family protein] |
0.583 |
0.069 |
| 58 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
0.582 |
-0.028 |
| 59 |
263593_at |
AT2G01860
|
EMB975, EMBRYO DEFECTIVE 975 |
0.582 |
0.365 |
| 60 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.581 |
-0.425 |
| 61 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
0.579 |
-0.017 |
| 62 |
266165_at |
AT2G28190
|
CSD2, copper/zinc superoxide dismutase 2, CZSOD2, COPPER/ZINC SUPEROXIDE DISMUTASE 2 |
0.578 |
0.136 |
| 63 |
254200_at |
AT4G24110
|
unknown |
0.575 |
0.333 |
| 64 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.572 |
0.317 |
| 65 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
0.566 |
0.555 |
| 66 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
0.559 |
0.295 |
| 67 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.558 |
0.762 |
| 68 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
0.550 |
0.184 |
| 69 |
259325_at |
AT3G05320
|
[O-fucosyltransferase family protein] |
0.547 |
0.523 |
| 70 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
0.546 |
0.335 |
| 71 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.544 |
0.635 |
| 72 |
267493_at |
AT2G30400
|
ATOFP2, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 2, OFP2, ovate family protein 2 |
0.537 |
0.057 |
| 73 |
264685_at |
AT1G65610
|
ATGH9A2, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9A2, KOR2, KORRIGAN 2 |
0.536 |
-0.013 |
| 74 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.535 |
0.028 |
| 75 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.530 |
0.544 |
| 76 |
263118_at |
AT1G03090
|
MCCA |
0.528 |
0.162 |
| 77 |
246790_at |
AT5G27610
|
ALY1, ALWAYS EARLY 1, ATALY1, ARABIDOPSIS THALIANA ALWAYS EARLY 1 |
0.526 |
0.048 |
| 78 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.526 |
0.315 |
| 79 |
249765_at |
AT5G24030
|
SLAH3, SLAC1 homologue 3 |
0.525 |
0.430 |
| 80 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.524 |
-0.038 |
| 81 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.522 |
0.485 |
| 82 |
249504_at |
AT5G38850
|
[Disease resistance protein (TIR-NBS-LRR class)] |
0.520 |
0.247 |
| 83 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
0.519 |
-0.063 |
| 84 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.518 |
-0.014 |
| 85 |
265147_at |
AT1G51380
|
[DEA(D/H)-box RNA helicase family protein] |
0.516 |
0.025 |
| 86 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
0.516 |
-0.453 |
| 87 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
0.515 |
-0.017 |
| 88 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.513 |
0.375 |
| 89 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.510 |
0.030 |
| 90 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.508 |
0.088 |
| 91 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
0.504 |
-0.014 |
| 92 |
247532_at |
AT5G61560
|
[U-box domain-containing protein kinase family protein] |
0.496 |
0.404 |
| 93 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.493 |
0.820 |
| 94 |
264809_at |
AT1G08830
|
CSD1, copper/zinc superoxide dismutase 1 |
0.484 |
0.101 |
| 95 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
0.482 |
0.061 |
| 96 |
258170_at |
AT3G21600
|
[Senescence/dehydration-associated protein-related] |
0.481 |
-0.053 |
| 97 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
0.479 |
0.090 |
| 98 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
0.477 |
0.278 |
| 99 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.476 |
0.166 |
| 100 |
254408_at |
AT4G21390
|
B120 |
0.475 |
0.620 |
| 101 |
266156_at |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
0.472 |
-0.095 |
| 102 |
248911_at |
AT5G45830
|
ATDOG1, DOG1, DELAY OF GERMINATION 1, GAAS5, germination ability after storage 5, GSQ5, GLUCOSE SENSING QTL 5 |
0.466 |
-0.034 |
| 103 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.464 |
-0.380 |
| 104 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
0.463 |
0.751 |
| 105 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.459 |
-0.012 |
| 106 |
265536_at |
AT2G15880
|
[Leucine-rich repeat (LRR) family protein] |
0.456 |
0.534 |
| 107 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
0.454 |
-0.491 |
| 108 |
265086_at |
AT1G03770
|
ATRING1B, ARABIDOPSIS THALIANA RING 1B, RING1B, RING 1B |
0.454 |
0.003 |
| 109 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
0.443 |
-0.335 |
| 110 |
245076_at |
AT2G23170
|
GH3.3 |
0.442 |
-0.017 |
| 111 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.442 |
0.053 |
| 112 |
258468_at |
AT3G06070
|
unknown |
0.441 |
0.724 |
| 113 |
263157_at |
AT1G54100
|
ALDH7B4, aldehyde dehydrogenase 7B4 |
0.436 |
-0.157 |
| 114 |
257643_at |
AT3G25730
|
EDF3, ethylene response DNA binding factor 3 |
0.427 |
0.119 |
| 115 |
262942_at |
AT1G79450
|
ALIS5, ALA-interacting subunit 5 |
0.420 |
0.055 |
| 116 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
0.419 |
0.253 |
| 117 |
247492_at |
AT5G61890
|
[Integrase-type DNA-binding superfamily protein] |
0.416 |
-0.111 |
| 118 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
0.411 |
-0.032 |
| 119 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
0.407 |
0.464 |
| 120 |
260048_at |
AT1G73750
|
[Uncharacterised conserved protein UCP031088, alpha/beta hydrolase] |
0.407 |
-0.124 |
| 121 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.406 |
0.529 |
| 122 |
261544_at |
AT1G63540
|
[hydroxyproline-rich glycoprotein family protein] |
0.405 |
0.004 |
| 123 |
262456_at |
AT1G11260
|
ATSTP1, SUGAR TRANSPORTER 1, STP1, sugar transporter 1 |
0.404 |
0.537 |
| 124 |
266231_at |
AT2G02220
|
ATPSKR1, PHYTOSULFOKIN RECEPTOR 1, PSKR1, phytosulfokin receptor 1 |
0.402 |
0.355 |
| 125 |
265276_at |
AT2G28400
|
unknown |
0.400 |
0.547 |
| 126 |
265999_at |
AT2G24100
|
ASG1, ALTERED SEED GERMINATION 1 |
0.400 |
-0.400 |
| 127 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
0.397 |
0.027 |
| 128 |
261934_at |
AT1G22400
|
ATUGT85A1, ARABIDOPSIS THALIANA UDP-GLUCOSYL TRANSFERASE 85A1, UGT85A1 |
0.395 |
0.346 |
| 129 |
260921_at |
AT1G21540
|
[AMP-dependent synthetase and ligase family protein] |
0.394 |
0.110 |
| 130 |
254707_at |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
0.393 |
0.378 |
| 131 |
263570_at |
AT2G27150
|
AAO3, abscisic aldehyde oxidase 3, AOdelta, Aldehyde oxidase delta, At-AO3, Arabidopsis thaliana aldehyde oxidase 3, AtAAO3 |
0.393 |
-0.158 |
| 132 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
0.391 |
0.193 |
| 133 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
0.391 |
0.353 |
| 134 |
253125_at |
AT4G36040
|
DJC23, DNA J protein C23, J11, DnaJ11 |
0.390 |
0.075 |
| 135 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
0.388 |
0.198 |
| 136 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
0.386 |
0.237 |
| 137 |
247410_at |
AT5G62990
|
emb1692, embryo defective 1692 |
0.386 |
0.237 |
| 138 |
250415_at |
AT5G11210
|
ATGLR2.5, ARABIDOPSIS THALIANA GLU, GLR2.5, glutamate receptor 2.5 |
0.384 |
0.029 |
| 139 |
256818_at |
AT3G21420
|
LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1 |
0.384 |
-0.061 |
| 140 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.383 |
0.212 |
| 141 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.380 |
0.154 |
| 142 |
263380_at |
AT2G40200
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.379 |
-0.048 |
| 143 |
246082_at |
AT5G20480
|
EFR, EF-TU receptor |
0.379 |
0.122 |
| 144 |
249136_at |
AT5G43180
|
unknown |
0.377 |
-0.221 |
| 145 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.377 |
0.375 |
| 146 |
245228_at |
AT3G29810
|
COBL2, COBRA-like protein 2 precursor |
0.376 |
0.114 |
| 147 |
259952_at |
AT1G71400
|
AtRLP12, receptor like protein 12, RLP12, receptor like protein 12 |
0.374 |
0.104 |
| 148 |
257654_at |
AT3G13310
|
DJC66, DNA J protein C66 |
0.374 |
0.277 |
| 149 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
0.373 |
0.026 |
| 150 |
266782_at |
AT2G29120
|
ATGLR2.7, glutamate receptor 2.7, GLR2.7, GLUTAMATE RECEPTOR 2.7, GLR2.7, glutamate receptor 2.7 |
0.370 |
-0.144 |
| 151 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.364 |
0.790 |
| 152 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
0.360 |
-0.042 |
| 153 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
0.357 |
0.003 |
| 154 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.356 |
0.180 |
| 155 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
0.356 |
0.172 |
| 156 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.354 |
-0.354 |
| 157 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.354 |
0.356 |
| 158 |
260581_at |
AT2G47190
|
ATMYB2, MYB DOMAIN PROTEIN 2, MYB2, myb domain protein 2 |
0.353 |
-0.106 |
| 159 |
246197_at |
AT4G37010
|
CEN2, centrin 2 |
0.352 |
0.003 |
| 160 |
264238_at |
AT1G54740
|
unknown |
0.350 |
0.867 |
| 161 |
259579_at |
AT1G28010
|
ABCB14, ATP-binding cassette B14, ATABCB14, Arabidopsis thaliana ATP-binding cassette B14, MDR12, multi-drug resistance 12, PGP14, P-glycoprotein 14 |
0.348 |
0.011 |
| 162 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.348 |
0.067 |
| 163 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.347 |
-0.814 |
| 164 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
0.346 |
-0.398 |
| 165 |
248239_at |
AT5G53920
|
[ribosomal protein L11 methyltransferase-related] |
0.346 |
0.109 |
| 166 |
264756_at |
AT1G61370
|
[S-locus lectin protein kinase family protein] |
0.345 |
0.082 |
| 167 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
0.344 |
0.041 |
| 168 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
0.344 |
-0.445 |
| 169 |
260337_at |
AT1G69310
|
ATWRKY57, WRKY57, WRKY DNA-binding protein 57 |
0.344 |
-0.200 |
| 170 |
256458_at |
AT1G75220
|
AtERDL6, ERDL6, ERD6-like 6 |
0.343 |
0.154 |
| 171 |
264767_at |
AT1G61380
|
SD1-29, S-domain-1 29 |
0.340 |
0.151 |
| 172 |
247444_at |
AT5G62630
|
HIPL2, hipl2 protein precursor |
0.340 |
0.061 |
| 173 |
246912_at |
AT5G25820
|
[Exostosin family protein] |
0.339 |
-0.047 |
| 174 |
265955_at |
AT2G37280
|
ABCG33, ATP-binding cassette G33, ATPDR5, PLEIOTROPIC DRUG RESISTANCE 5, PDR5, pleiotropic drug resistance 5 |
0.337 |
0.003 |
| 175 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.335 |
0.605 |
| 176 |
261674_at |
AT1G18270
|
[ketose-bisphosphate aldolase class-II family protein] |
0.334 |
-0.010 |
| 177 |
264082_at |
AT2G28570
|
unknown |
0.334 |
0.007 |
| 178 |
263702_at |
AT1G31240
|
[Bromodomain transcription factor] |
0.334 |
-0.096 |
| 179 |
253175_at |
AT4G35050
|
MSI3, MULTICOPY SUPPRESSOR OF IRA1 3, NFC3, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 3 |
0.333 |
0.313 |
| 180 |
258764_at |
AT3G10720
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.332 |
0.371 |
| 181 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.332 |
0.477 |
| 182 |
256525_at |
AT1G66180
|
[Eukaryotic aspartyl protease family protein] |
0.332 |
0.348 |
| 183 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.331 |
-0.248 |
| 184 |
251914_at |
AT3G53930
|
[Protein kinase superfamily protein] |
0.331 |
0.159 |
| 185 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.328 |
0.262 |
| 186 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.328 |
0.993 |
| 187 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
0.328 |
0.403 |
| 188 |
248763_at |
AT5G47550
|
[Cystatin/monellin superfamily protein] |
0.328 |
-0.114 |
| 189 |
262228_at |
AT1G68690
|
AtPERK9, proline-rich extensin-like receptor kinase 9, PERK9, proline-rich extensin-like receptor kinase 9 |
0.328 |
0.241 |
| 190 |
266106_at |
AT2G45170
|
ATATG8E, AUTOPHAGY 8E, ATG8E, AUTOPHAGY 8E |
0.327 |
0.499 |
| 191 |
256720_at |
AT2G34140
|
[Dof-type zinc finger DNA-binding family protein] |
0.327 |
0.250 |
| 192 |
257479_at |
AT1G16150
|
WAKL4, wall associated kinase-like 4 |
0.326 |
0.008 |
| 193 |
252289_at |
AT3G49130
|
[SWAP (Suppressor-of-White-APricot)/surp RNA-binding domain-containing protein] |
0.325 |
-0.008 |
| 194 |
265481_at |
AT2G15960
|
unknown |
0.325 |
0.313 |
| 195 |
267624_at |
AT2G39660
|
BIK1, botrytis-induced kinase1 |
0.324 |
0.133 |
| 196 |
253458_at |
AT4G32070
|
Phox4, Phox4 |
0.324 |
-0.102 |
| 197 |
262844_at |
AT1G14890
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.324 |
0.080 |
| 198 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
0.322 |
0.577 |
| 199 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
0.321 |
-0.236 |
| 200 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.320 |
-0.729 |
| 201 |
255905_at |
AT1G17810
|
BETA-TIP, beta-tonoplast intrinsic protein |
0.317 |
-0.031 |
| 202 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.317 |
0.539 |
| 203 |
261991_at |
AT1G33700
|
[Beta-glucosidase, GBA2 type family protein] |
0.317 |
0.008 |
| 204 |
258282_at |
AT3G26910
|
[hydroxyproline-rich glycoprotein family protein] |
0.316 |
0.370 |
| 205 |
265028_at |
AT1G24530
|
[Transducin/WD40 repeat-like superfamily protein] |
0.315 |
0.433 |
| 206 |
249139_at |
AT5G43170
|
AZF3, zinc-finger protein 3, ZF3, zinc-finger protein 3 |
0.313 |
0.083 |
| 207 |
265680_at |
AT2G32150
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.312 |
0.019 |
| 208 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
0.312 |
0.513 |
| 209 |
253019_at |
AT4G38010
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.309 |
0.119 |
| 210 |
252100_at |
AT3G51110
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.308 |
0.032 |
| 211 |
246999_at |
AT5G67440
|
MEL2, MAB4/ENP/NPY1-LIKE 2, NPY3, NAKED PINS IN YUC MUTANTS 3 |
0.307 |
0.041 |
| 212 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-1.349 |
-0.631 |
| 213 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-1.216 |
-0.058 |
| 214 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-1.194 |
-0.109 |
| 215 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-1.192 |
-0.239 |
| 216 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-1.179 |
-0.015 |
| 217 |
252888_at |
AT4G39210
|
APL3 |
-1.080 |
-0.274 |
| 218 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-1.030 |
-0.650 |
| 219 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-1.000 |
0.098 |
| 220 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.996 |
-0.099 |
| 221 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.980 |
0.547 |
| 222 |
266132_at |
AT2G45130
|
ATSPX3, ARABIDOPSIS THALIANA SPX DOMAIN GENE 3, SPX3, SPX domain gene 3 |
-0.977 |
-0.006 |
| 223 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
-0.967 |
0.040 |
| 224 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.936 |
-0.329 |
| 225 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.934 |
-0.443 |
| 226 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.926 |
-0.262 |
| 227 |
259399_at |
AT1G17710
|
AtPEPC1, Arabidopsis thaliana phosphoethanolamine/phosphocholine phosphatase 1, PEPC1, phosphoethanolamine/phosphocholine phosphatase 1 |
-0.895 |
0.000 |
| 228 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.893 |
-0.198 |
| 229 |
249004_at |
AT5G44570
|
unknown |
-0.889 |
0.104 |
| 230 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.880 |
-0.122 |
| 231 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.868 |
-0.010 |
| 232 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.865 |
0.132 |
| 233 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
-0.862 |
0.025 |
| 234 |
246001_at |
AT5G20790
|
unknown |
-0.851 |
0.118 |
| 235 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
-0.846 |
0.086 |
| 236 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
-0.830 |
-0.481 |
| 237 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.815 |
-0.379 |
| 238 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
-0.810 |
-0.354 |
| 239 |
248867_at |
AT5G46830
|
ATNIG1, NACL-INDUCIBLE GENE 1, NIG1, NACL-inducible gene 1 |
-0.809 |
-0.809 |
| 240 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
-0.809 |
-0.109 |
| 241 |
262651_at |
AT1G14100
|
FUT8, fucosyltransferase 8 |
-0.799 |
0.050 |
| 242 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.795 |
-0.442 |
| 243 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.793 |
0.067 |
| 244 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
-0.782 |
0.044 |
| 245 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.772 |
-0.294 |
| 246 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.769 |
0.262 |
| 247 |
253940_at |
AT4G26950
|
unknown |
-0.766 |
-0.288 |
| 248 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
-0.764 |
-0.088 |
| 249 |
263031_at |
AT1G24070
|
ATCSLA10, cellulose synthase-like A10, CSLA10, CELLULOSE SYNTHASE LIKE A10, CSLA10, cellulose synthase-like A10 |
-0.763 |
-0.748 |
| 250 |
261814_at |
AT1G08310
|
[alpha/beta-Hydrolases superfamily protein] |
-0.760 |
-0.148 |
| 251 |
257421_at |
AT1G12030
|
unknown |
-0.758 |
0.246 |
| 252 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.756 |
-0.315 |
| 253 |
261921_at |
AT1G65900
|
unknown |
-0.742 |
-0.107 |
| 254 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.733 |
-0.778 |
| 255 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
-0.719 |
-0.300 |
| 256 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.715 |
-0.265 |
| 257 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.697 |
0.713 |
| 258 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
-0.694 |
-0.091 |
| 259 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.692 |
-0.312 |
| 260 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.689 |
-0.373 |
| 261 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.684 |
-0.166 |
| 262 |
266766_at |
AT2G46880
|
ATPAP14, PAP14, purple acid phosphatase 14 |
-0.679 |
0.105 |
| 263 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-0.678 |
-0.645 |
| 264 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
-0.677 |
-0.060 |
| 265 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.676 |
-0.242 |
| 266 |
256924_at |
AT3G29590
|
AT5MAT |
-0.676 |
-0.606 |
| 267 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.673 |
0.078 |
| 268 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.657 |
-0.687 |
| 269 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.652 |
0.224 |
| 270 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
-0.637 |
-0.453 |
| 271 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
-0.631 |
-0.345 |
| 272 |
254290_at |
AT4G23000
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.631 |
-0.145 |
| 273 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.628 |
-0.087 |
| 274 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.620 |
-0.246 |
| 275 |
253317_at |
AT4G33960
|
unknown |
-0.618 |
0.282 |
| 276 |
262700_at |
AT1G76020
|
[Thioredoxin superfamily protein] |
-0.618 |
-0.003 |
| 277 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.616 |
0.133 |
| 278 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.615 |
-0.210 |
| 279 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.611 |
-0.084 |
| 280 |
265305_at |
AT2G20340
|
AAS, aromatic aldehyde synthase, AtAAS |
-0.610 |
-0.148 |
| 281 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.610 |
-0.407 |
| 282 |
267555_at |
AT2G32765
|
ATSUMO5, SUM5, SUMO 5, SUMO5, small ubiquitinrelated modifier 5 |
-0.600 |
0.004 |
| 283 |
262236_at |
AT1G48330
|
unknown |
-0.599 |
-0.362 |
| 284 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.599 |
0.043 |
| 285 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
-0.599 |
0.223 |
| 286 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
-0.590 |
0.363 |
| 287 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-0.588 |
-0.039 |
| 288 |
251143_at |
AT5G01220
|
SQD2, sulfoquinovosyldiacylglycerol 2 |
-0.586 |
0.055 |
| 289 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
-0.584 |
0.196 |
| 290 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.581 |
0.128 |
| 291 |
266738_at |
AT2G47010
|
unknown |
-0.578 |
-0.044 |
| 292 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
-0.576 |
-0.235 |
| 293 |
254632_at |
AT4G18630
|
unknown |
-0.574 |
-0.235 |
| 294 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.574 |
-0.260 |
| 295 |
253947_at |
AT4G26760
|
MAP65-2, microtubule-associated protein 65-2 |
-0.573 |
0.042 |
| 296 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.566 |
0.235 |
| 297 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
-0.565 |
0.212 |
| 298 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
-0.563 |
-0.276 |
| 299 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.562 |
-0.195 |
| 300 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.562 |
0.059 |
| 301 |
249495_at |
AT5G39100
|
GLP6, germin-like protein 6 |
-0.560 |
-0.079 |
| 302 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.560 |
0.035 |
| 303 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.559 |
-0.174 |
| 304 |
256441_at |
AT3G10940
|
LSF2, LIKE SEX4 2 |
-0.551 |
0.017 |
| 305 |
249002_at |
AT5G44520
|
[NagB/RpiA/CoA transferase-like superfamily protein] |
-0.550 |
-0.034 |
| 306 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
-0.549 |
-0.070 |
| 307 |
255460_at |
AT4G02800
|
unknown |
-0.544 |
0.098 |
| 308 |
252365_at |
AT3G48350
|
CEP3, cysteine endopeptidase 3 |
-0.542 |
0.064 |
| 309 |
260262_at |
AT1G68470
|
[Exostosin family protein] |
-0.542 |
-0.147 |
| 310 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.540 |
-0.815 |
| 311 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.539 |
0.099 |
| 312 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.539 |
-0.127 |
| 313 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
-0.535 |
0.185 |
| 314 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.523 |
-0.455 |
| 315 |
266655_at |
AT2G25880
|
AtAUR2, ataurora2, AUR2, ataurora2 |
-0.517 |
0.090 |
| 316 |
247041_at |
AT5G67180
|
TOE3, target of early activation tagged (EAT) 3 |
-0.516 |
-0.158 |
| 317 |
250696_at |
AT5G06790
|
unknown |
-0.512 |
-0.358 |
| 318 |
245348_at |
AT4G17770
|
ATTPS5, trehalose phosphatase/synthase 5, TPS5, TREHALOSE -6-PHOSPHATASE SYNTHASE S5, TPS5, trehalose phosphatase/synthase 5 |
-0.509 |
-0.226 |
| 319 |
263597_at |
AT2G01870
|
unknown |
-0.508 |
-0.033 |
| 320 |
262411_at |
AT1G34640
|
[peptidases] |
-0.508 |
-0.245 |
| 321 |
245479_at |
AT4G16140
|
[proline-rich family protein] |
-0.507 |
-0.109 |
| 322 |
265277_at |
AT2G28410
|
unknown |
-0.507 |
-0.128 |
| 323 |
261593_at |
AT1G33170
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.507 |
-0.092 |
| 324 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.505 |
-0.443 |
| 325 |
259996_at |
AT1G67910
|
unknown |
-0.505 |
0.546 |
| 326 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
-0.505 |
0.139 |
| 327 |
263902_at |
AT2G36230
|
APG10, ALBINO AND PALE GREEN 10, HISN3 |
-0.504 |
0.105 |
| 328 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.504 |
-0.446 |
| 329 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
-0.503 |
0.355 |
| 330 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.501 |
0.229 |
| 331 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.500 |
-0.049 |
| 332 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.498 |
-0.105 |
| 333 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
-0.496 |
-0.440 |
| 334 |
260666_at |
AT1G19300
|
ATGATL1, GATL1, GALACTURONOSYLTRANSFERASE-LIKE 1, GLZ1, GAOLAOZHUANGREN 1, PARVUS, PARVUS |
-0.494 |
0.025 |
| 335 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.492 |
0.046 |
| 336 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
-0.491 |
-0.200 |
| 337 |
253729_at |
AT4G29360
|
[O-Glycosyl hydrolases family 17 protein] |
-0.491 |
-0.226 |
| 338 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
-0.490 |
-0.245 |
| 339 |
247713_at |
AT5G59330
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.488 |
-0.091 |
| 340 |
245930_at |
AT5G09240
|
[ssDNA-binding transcriptional regulator] |
-0.486 |
0.121 |
| 341 |
261596_at |
AT1G33080
|
[MATE efflux family protein] |
-0.483 |
0.027 |
| 342 |
254485_at |
AT4G20760
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.483 |
0.008 |
| 343 |
249046_at |
AT5G44400
|
[FAD-binding Berberine family protein] |
-0.481 |
0.065 |
| 344 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.480 |
-0.095 |
| 345 |
256860_at |
AT3G23840
|
CER26-LIKE, CER26-LIKE |
-0.479 |
-0.189 |
| 346 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
-0.477 |
0.018 |
| 347 |
257191_at |
AT3G13175
|
unknown |
-0.477 |
0.135 |
| 348 |
256585_at |
AT3G28760
|
unknown |
-0.475 |
-0.006 |
| 349 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.474 |
-0.093 |
| 350 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.473 |
-0.017 |
| 351 |
257875_at |
AT3G17120
|
unknown |
-0.472 |
-0.292 |
| 352 |
261221_at |
AT1G19960
|
unknown |
-0.467 |
-0.008 |
| 353 |
252089_at |
AT3G52110
|
unknown |
-0.467 |
0.181 |
| 354 |
259042_at |
AT3G03450
|
RGL2, RGA-like 2 |
-0.466 |
-0.572 |
| 355 |
257750_at |
AT3G18800
|
unknown |
-0.466 |
-0.133 |
| 356 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.465 |
-0.622 |
| 357 |
255664_at |
AT4G00440
|
TRM15, TON1 Recruiting Motif 15 |
-0.463 |
-0.049 |
| 358 |
254119_at |
AT4G24780
|
[Pectin lyase-like superfamily protein] |
-0.463 |
-0.336 |
| 359 |
260041_at |
AT1G68780
|
[RNI-like superfamily protein] |
-0.457 |
-0.220 |
| 360 |
258264_at |
AT3G15790
|
ATMBD11, MBD11, methyl-CPG-binding domain 11 |
-0.456 |
-0.199 |
| 361 |
259014_at |
AT3G07320
|
[O-Glycosyl hydrolases family 17 protein] |
-0.454 |
0.312 |
| 362 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
-0.453 |
-0.046 |
| 363 |
248057_at |
AT5G55520
|
unknown |
-0.452 |
0.103 |
| 364 |
255488_at |
AT4G02630
|
[Protein kinase superfamily protein] |
-0.451 |
-0.155 |
| 365 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
-0.449 |
-0.067 |
| 366 |
254693_at |
AT4G17880
|
MYC4 |
-0.448 |
-0.585 |
| 367 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.447 |
0.006 |
| 368 |
255924_at |
AT1G22170
|
[Phosphoglycerate mutase family protein] |
-0.447 |
0.030 |
| 369 |
266413_at |
AT2G38740
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.447 |
-0.151 |
| 370 |
252248_at |
AT3G49750
|
AtRLP44, receptor like protein 44, RLP44, receptor like protein 44 |
-0.447 |
-0.073 |
| 371 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.446 |
-0.093 |
| 372 |
249718_at |
AT5G35740
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.444 |
0.085 |
| 373 |
251493_at |
AT3G59300
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.444 |
-0.159 |
| 374 |
248596_at |
AT5G49330
|
ATMYB111, ARABIDOPSIS MYB DOMAIN PROTEIN 111, MYB111, myb domain protein 111, PFG3, PRODUCTION OF FLAVONOL GLYCOSIDES 3 |
-0.443 |
-0.051 |
| 375 |
257663_at |
AT3G20260
|
[FUNCTIONS IN: structural constituent of ribosome] |
-0.440 |
-0.057 |
| 376 |
254633_at |
AT4G18640
|
MRH1, morphogenesis of root hair 1 |
-0.440 |
0.015 |
| 377 |
264802_at |
AT1G08560
|
ATSYP111, KN, KNOLLE, SYP111, syntaxin of plants 111 |
-0.440 |
0.263 |
| 378 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
-0.439 |
-0.260 |
| 379 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
-0.438 |
-0.583 |
| 380 |
253337_at |
AT4G33470
|
ATHDA14, HDA14, histone deacetylase 14 |
-0.438 |
-0.008 |
| 381 |
245133_at |
AT2G45310
|
GAE4, UDP-D-glucuronate 4-epimerase 4 |
-0.436 |
-0.152 |
| 382 |
265339_at |
AT2G18230
|
AtPPa2, pyrophosphorylase 2, PPa2, pyrophosphorylase 2 |
-0.436 |
-0.212 |
| 383 |
258339_at |
AT3G16120
|
[Dynein light chain type 1 family protein] |
-0.434 |
-0.020 |
| 384 |
251319_at |
AT3G61610
|
[Galactose mutarotase-like superfamily protein] |
-0.430 |
-0.078 |
| 385 |
265454_at |
AT2G46530
|
ARF11, auxin response factor 11 |
-0.430 |
-0.027 |
| 386 |
262313_at |
AT1G70900
|
unknown |
-0.430 |
-0.165 |
| 387 |
251395_at |
AT2G45470
|
AGP8, ARABINOGALACTAN PROTEIN 8, FLA8, FASCICLIN-like arabinogalactan protein 8 |
-0.429 |
0.020 |
| 388 |
246899_at |
AT5G25590
|
[INVOLVED IN: N-terminal protein myristoylation] |
-0.428 |
-0.161 |
| 389 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.428 |
-0.448 |
| 390 |
256675_at |
AT3G52170
|
[DNA binding] |
-0.427 |
0.282 |
| 391 |
251982_at |
AT3G53190
|
[Pectin lyase-like superfamily protein] |
-0.427 |
0.057 |
| 392 |
262134_at |
AT1G77990
|
AST56, SULTR2;2, SULPHATE TRANSPORTER 2;2 |
-0.425 |
0.154 |
| 393 |
264839_at |
AT1G03630
|
POR C, protochlorophyllide oxidoreductase C, PORC |
-0.425 |
-0.028 |
| 394 |
252077_at |
AT3G51720
|
unknown |
-0.425 |
0.065 |
| 395 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
-0.425 |
0.044 |
| 396 |
258376_at |
AT3G17680
|
[Kinase interacting (KIP1-like) family protein] |
-0.424 |
0.103 |
| 397 |
255727_at |
AT1G25510
|
[Eukaryotic aspartyl protease family protein] |
-0.423 |
-0.045 |
| 398 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.421 |
-0.015 |
| 399 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
-0.421 |
0.057 |
| 400 |
249588_at |
AT5G37790
|
[Protein kinase superfamily protein] |
-0.419 |
-0.117 |
| 401 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
-0.418 |
0.170 |
| 402 |
253296_at |
AT4G33770
|
ITPK2, inositol 1,3,4-trisphosphate 5/6 kinase 2 |
-0.417 |
-0.145 |
| 403 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.416 |
-0.025 |
| 404 |
266170_at |
AT2G39050
|
ArathEULS3, EULS3, Euonymus lectin S3 |
-0.416 |
-0.094 |
| 405 |
260801_at |
AT1G78430
|
RIP4, ROP interactive partner 4 |
-0.415 |
0.046 |
| 406 |
267429_at |
AT2G34850
|
MEE25, maternal effect embryo arrest 25 |
-0.414 |
-0.011 |
| 407 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.412 |
-0.247 |
| 408 |
261769_at |
AT1G76100
|
PETE1, plastocyanin 1 |
-0.411 |
0.004 |
| 409 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.410 |
0.116 |
| 410 |
258932_at |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
-0.410 |
-0.299 |
| 411 |
247097_at |
AT5G66460
|
AtMAN7, MAN7, endo-beta-mannase 7 |
-0.408 |
-0.263 |
| 412 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.408 |
-0.276 |
| 413 |
261025_at |
AT1G01225
|
[NC domain-containing protein-related] |
-0.408 |
0.025 |
| 414 |
253165_at |
AT4G35320
|
unknown |
-0.407 |
0.044 |
| 415 |
263687_at |
AT1G26940
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.406 |
0.017 |
| 416 |
258471_at |
AT3G06030
|
ANP3, NPK1-related protein kinase 3, AtANP3, MAPKKK12, NP3, NPK1-related protein kinase 3 |
-0.406 |
0.142 |
| 417 |
264907_at |
AT2G17280
|
[Phosphoglycerate mutase family protein] |
-0.405 |
0.225 |
| 418 |
261536_at |
AT1G01790
|
ATKEA1, K+ EFFLUX ANTIPORTER 1, KEA1, K+ efflux antiporter 1 |
-0.404 |
-0.074 |
| 419 |
262181_at |
AT1G78060
|
[Glycosyl hydrolase family protein] |
-0.401 |
0.022 |
| 420 |
265994_at |
AT2G24170
|
[Endomembrane protein 70 protein family] |
-0.400 |
-0.073 |
| 421 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
-0.399 |
-0.286 |