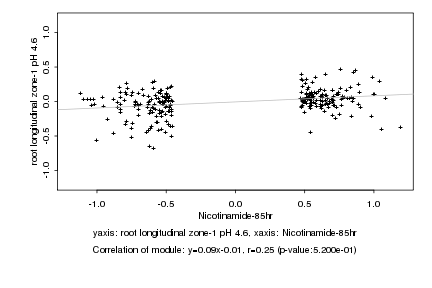
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Nicotinamide-85hr |
Log2 signal ratio
root longitudinal zone-1 pH 4.6 |
| 1 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
1.189 |
-0.365 |
| 2 |
260664_at |
AT1G19510
|
ATRL5, RAD-like 5, RL5, RAD-like 5, RSM4, RADIALIS-LIKE SANT/MYB 4 |
1.077 |
0.049 |
| 3 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
1.054 |
-0.394 |
| 4 |
248563_at |
AT5G49690
|
[UDP-Glycosyltransferase superfamily protein] |
1.036 |
0.292 |
| 5 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
1.003 |
0.110 |
| 6 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.993 |
0.116 |
| 7 |
261475_at |
AT1G14550
|
[Peroxidase superfamily protein] |
0.988 |
0.360 |
| 8 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.978 |
-0.205 |
| 9 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.901 |
-0.086 |
| 10 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.894 |
0.133 |
| 11 |
265892_at |
AT2G15020
|
unknown |
0.885 |
-0.037 |
| 12 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.884 |
0.255 |
| 13 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.861 |
0.451 |
| 14 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.851 |
0.050 |
| 15 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.846 |
0.423 |
| 16 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.839 |
0.005 |
| 17 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.832 |
0.072 |
| 18 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.832 |
-0.203 |
| 19 |
256442_at |
AT3G10930
|
unknown |
0.824 |
0.216 |
| 20 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
0.819 |
0.046 |
| 21 |
245705_at |
AT5G04390
|
[C2H2-type zinc finger family protein] |
0.807 |
0.050 |
| 22 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
0.801 |
0.165 |
| 23 |
250098_at |
AT5G17350
|
unknown |
0.781 |
0.068 |
| 24 |
259953_at |
AT1G74810
|
BOR5, REQUIRES HIGH BORON 5 |
0.773 |
0.082 |
| 25 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.769 |
0.049 |
| 26 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.761 |
0.031 |
| 27 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
0.760 |
-0.054 |
| 28 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
0.757 |
0.189 |
| 29 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.756 |
0.465 |
| 30 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.747 |
-0.174 |
| 31 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.739 |
0.105 |
| 32 |
263475_at |
AT2G31945
|
unknown |
0.737 |
0.132 |
| 33 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
0.733 |
0.106 |
| 34 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.728 |
-0.076 |
| 35 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.724 |
0.102 |
| 36 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
0.721 |
-0.237 |
| 37 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.713 |
-0.047 |
| 38 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.707 |
-0.048 |
| 39 |
258752_at |
AT3G09520
|
ATEXO70H4, exocyst subunit exo70 family protein H4, EXO70H4, exocyst subunit exo70 family protein H4 |
0.704 |
0.018 |
| 40 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
0.704 |
0.076 |
| 41 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.700 |
0.170 |
| 42 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.698 |
0.043 |
| 43 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.697 |
-0.191 |
| 44 |
259511_at |
AT1G12520
|
ATCCS, copper chaperone for SOD1, CCS, copper chaperone for SOD1 |
0.697 |
-0.012 |
| 45 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.696 |
0.166 |
| 46 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
0.690 |
0.038 |
| 47 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
0.679 |
-0.022 |
| 48 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.673 |
0.010 |
| 49 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.670 |
-0.076 |
| 50 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.663 |
0.054 |
| 51 |
249798_at |
AT5G23730
|
EFO2, EARLY FLOWERING BY OVEREXPRESSION 2, RUP2, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 2 |
0.660 |
-0.034 |
| 52 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.656 |
-0.028 |
| 53 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
0.655 |
0.100 |
| 54 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.650 |
0.021 |
| 55 |
253643_at |
AT4G29780
|
unknown |
0.648 |
-0.036 |
| 56 |
249454_at |
AT5G39520
|
unknown |
0.647 |
0.092 |
| 57 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
0.646 |
0.405 |
| 58 |
260744_at |
AT1G15010
|
unknown |
0.637 |
0.171 |
| 59 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.636 |
-0.142 |
| 60 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
0.628 |
0.114 |
| 61 |
265203_at |
AT2G36630
|
[Sulfite exporter TauE/SafE family protein] |
0.622 |
0.011 |
| 62 |
266165_at |
AT2G28190
|
CSD2, copper/zinc superoxide dismutase 2, CZSOD2, COPPER/ZINC SUPEROXIDE DISMUTASE 2 |
0.622 |
-0.047 |
| 63 |
266247_at |
AT2G27660
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.622 |
0.070 |
| 64 |
265276_at |
AT2G28400
|
unknown |
0.618 |
0.073 |
| 65 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
0.617 |
-0.088 |
| 66 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
0.616 |
0.081 |
| 67 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.613 |
0.029 |
| 68 |
247205_at |
AT5G64890
|
PROPEP2, elicitor peptide 2 precursor |
0.613 |
-0.041 |
| 69 |
245228_at |
AT3G29810
|
COBL2, COBRA-like protein 2 precursor |
0.613 |
0.185 |
| 70 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.611 |
0.083 |
| 71 |
246485_at |
AT5G16080
|
AtCXE17, carboxyesterase 17, CXE17, carboxyesterase 17 |
0.608 |
-0.059 |
| 72 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
0.608 |
0.022 |
| 73 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.596 |
0.159 |
| 74 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
0.582 |
-0.071 |
| 75 |
260581_at |
AT2G47190
|
ATMYB2, MYB DOMAIN PROTEIN 2, MYB2, myb domain protein 2 |
0.581 |
0.117 |
| 76 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
0.580 |
0.026 |
| 77 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.571 |
0.355 |
| 78 |
260337_at |
AT1G69310
|
ATWRKY57, WRKY57, WRKY DNA-binding protein 57 |
0.571 |
-0.027 |
| 79 |
254710_at |
AT4G18050
|
ABCB9, ATP-binding cassette B9, PGP9, P-glycoprotein 9 |
0.564 |
0.143 |
| 80 |
264809_at |
AT1G08830
|
CSD1, copper/zinc superoxide dismutase 1 |
0.561 |
-0.013 |
| 81 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.559 |
0.080 |
| 82 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
0.558 |
0.050 |
| 83 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.554 |
0.282 |
| 84 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.552 |
0.064 |
| 85 |
263196_at |
AT1G36070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.552 |
0.113 |
| 86 |
253814_at |
AT4G28290
|
unknown |
0.548 |
0.092 |
| 87 |
261544_at |
AT1G63540
|
[hydroxyproline-rich glycoprotein family protein] |
0.546 |
0.053 |
| 88 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.546 |
0.112 |
| 89 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.543 |
-0.011 |
| 90 |
258275_at |
AT3G15760
|
unknown |
0.543 |
-0.045 |
| 91 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.543 |
0.138 |
| 92 |
259982_at |
AT1G76410
|
ATL8 |
0.541 |
-0.018 |
| 93 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.541 |
0.016 |
| 94 |
259856_at |
AT1G68440
|
unknown |
0.539 |
-0.093 |
| 95 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
0.537 |
-0.449 |
| 96 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.534 |
-0.071 |
| 97 |
248207_at |
AT5G53970
|
TAT7, tyrosine aminotransferase 7 |
0.534 |
0.100 |
| 98 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
0.532 |
0.060 |
| 99 |
258813_at |
AT3G04060
|
anac046, NAC domain containing protein 46, NAC046, NAC domain containing protein 46 |
0.531 |
0.041 |
| 100 |
254410_at |
AT4G21410
|
CRK29, cysteine-rich RLK (RECEPTOR-like protein kinase) 29 |
0.528 |
0.063 |
| 101 |
265084_at |
AT1G03790
|
AtTZF4, SOM, SOMNUS, TZF4, Tandem CCCH Zinc Finger protein 4 |
0.528 |
0.130 |
| 102 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.525 |
0.044 |
| 103 |
245420_at |
AT4G17410
|
[DWNN domain, a CCHC-type zinc finger] |
0.524 |
0.073 |
| 104 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
0.521 |
0.216 |
| 105 |
251621_at |
AT3G57700
|
[Protein kinase superfamily protein] |
0.519 |
0.179 |
| 106 |
257654_at |
AT3G13310
|
DJC66, DNA J protein C66 |
0.518 |
0.040 |
| 107 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
0.511 |
0.107 |
| 108 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
0.510 |
0.330 |
| 109 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
0.510 |
-0.019 |
| 110 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
0.507 |
0.070 |
| 111 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.507 |
0.122 |
| 112 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.502 |
0.274 |
| 113 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.501 |
0.041 |
| 114 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.499 |
0.009 |
| 115 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.498 |
0.030 |
| 116 |
248700_at |
AT5G48400
|
ATGLR1.2, GLR1.2, GLUTAMATE RECEPTOR 1.2 |
0.493 |
-0.010 |
| 117 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
0.493 |
-0.158 |
| 118 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.487 |
0.030 |
| 119 |
249765_at |
AT5G24030
|
SLAH3, SLAC1 homologue 3 |
0.485 |
-0.004 |
| 120 |
255926_at |
AT1G22190
|
RAP2.4, related to AP2 4 |
0.482 |
0.024 |
| 121 |
264685_at |
AT1G65610
|
ATGH9A2, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9A2, KOR2, KORRIGAN 2 |
0.482 |
0.218 |
| 122 |
258188_at |
AT3G17800
|
unknown |
0.482 |
0.042 |
| 123 |
259711_at |
AT1G77570
|
AtREN1, REN1, restricted to nucleolus 1 |
0.480 |
0.318 |
| 124 |
254063_at |
AT4G25390
|
[Protein kinase superfamily protein] |
0.479 |
0.008 |
| 125 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
0.478 |
-0.048 |
| 126 |
267066_at |
AT2G41040
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.475 |
0.135 |
| 127 |
251727_at |
AT3G56290
|
unknown |
0.475 |
0.326 |
| 128 |
245353_at |
AT4G16000
|
unknown |
0.474 |
-0.058 |
| 129 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
0.471 |
0.398 |
| 130 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
0.470 |
-0.080 |
| 131 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.469 |
0.055 |
| 132 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-1.125 |
0.124 |
| 133 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-1.102 |
0.039 |
| 134 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-1.073 |
0.032 |
| 135 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-1.048 |
0.041 |
| 136 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-1.046 |
-0.045 |
| 137 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-1.031 |
0.043 |
| 138 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-1.020 |
-0.038 |
| 139 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
-1.006 |
-0.552 |
| 140 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.967 |
0.068 |
| 141 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-0.957 |
-0.065 |
| 142 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
-0.924 |
-0.247 |
| 143 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.884 |
0.031 |
| 144 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
-0.883 |
-0.456 |
| 145 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.855 |
-0.003 |
| 146 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.853 |
-0.077 |
| 147 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.841 |
0.212 |
| 148 |
258080_at |
AT3G25930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.836 |
-0.032 |
| 149 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
-0.836 |
0.141 |
| 150 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
-0.836 |
-0.103 |
| 151 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.833 |
0.066 |
| 152 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.832 |
-0.148 |
| 153 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.802 |
0.029 |
| 154 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
-0.801 |
0.144 |
| 155 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-0.801 |
-0.330 |
| 156 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.792 |
0.110 |
| 157 |
265050_at |
AT1G52070
|
[Mannose-binding lectin superfamily protein] |
-0.788 |
-0.286 |
| 158 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
-0.787 |
0.264 |
| 159 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
-0.783 |
0.200 |
| 160 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
-0.755 |
-0.511 |
| 161 |
249545_at |
AT5G38030
|
[MATE efflux family protein] |
-0.755 |
0.101 |
| 162 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
-0.752 |
-0.380 |
| 163 |
253720_at |
AT4G29270
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.746 |
-0.307 |
| 164 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
-0.745 |
0.141 |
| 165 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.735 |
-0.063 |
| 166 |
266330_at |
AT2G01530
|
MLP329, MLP-like protein 329, ZCE2, (Zusammen-CA)-enhanced 2 |
-0.728 |
-0.041 |
| 167 |
250059_at |
AT5G17820
|
[Peroxidase superfamily protein] |
-0.725 |
-0.004 |
| 168 |
266132_at |
AT2G45130
|
ATSPX3, ARABIDOPSIS THALIANA SPX DOMAIN GENE 3, SPX3, SPX domain gene 3 |
-0.722 |
-0.031 |
| 169 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
-0.722 |
0.014 |
| 170 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.709 |
-0.035 |
| 171 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.704 |
-0.044 |
| 172 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.704 |
0.118 |
| 173 |
253947_at |
AT4G26760
|
MAP65-2, microtubule-associated protein 65-2 |
-0.703 |
-0.198 |
| 174 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.700 |
-0.110 |
| 175 |
256675_at |
AT3G52170
|
[DNA binding] |
-0.686 |
-0.032 |
| 176 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.673 |
0.184 |
| 177 |
253337_at |
AT4G33470
|
ATHDA14, HDA14, histone deacetylase 14 |
-0.665 |
0.098 |
| 178 |
246415_at |
AT5G17160
|
unknown |
-0.644 |
-0.446 |
| 179 |
255876_at |
AT2G40480
|
unknown |
-0.642 |
-0.085 |
| 180 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-0.634 |
-0.032 |
| 181 |
258341_at |
AT3G22790
|
NET1A, Networked 1A |
-0.634 |
-0.407 |
| 182 |
261806_at |
AT1G30510
|
ATRFNR2, root FNR 2, RFNR2, root FNR 2 |
-0.632 |
-0.002 |
| 183 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
-0.631 |
0.082 |
| 184 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.624 |
-0.652 |
| 185 |
252833_at |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
-0.623 |
-0.161 |
| 186 |
258333_at |
AT3G16000
|
MFP1, MAR binding filament-like protein 1 |
-0.621 |
0.011 |
| 187 |
261814_at |
AT1G08310
|
[alpha/beta-Hydrolases superfamily protein] |
-0.616 |
-0.123 |
| 188 |
265042_at |
AT1G04040
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.616 |
-0.379 |
| 189 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.611 |
0.037 |
| 190 |
253957_at |
AT4G26320
|
AGP13, arabinogalactan protein 13 |
-0.607 |
-0.356 |
| 191 |
259276_at |
AT3G01190
|
[Peroxidase superfamily protein] |
-0.604 |
-0.151 |
| 192 |
248150_at |
AT5G54670
|
ATK3, kinesin 3, KATC, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA C |
-0.601 |
-0.076 |
| 193 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-0.600 |
-0.155 |
| 194 |
248057_at |
AT5G55520
|
unknown |
-0.599 |
0.278 |
| 195 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.598 |
-0.101 |
| 196 |
263597_at |
AT2G01870
|
unknown |
-0.596 |
0.201 |
| 197 |
263352_at |
AT2G22080
|
unknown |
-0.593 |
-0.669 |
| 198 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.589 |
-0.042 |
| 199 |
254333_at |
AT4G22753
|
ATSMO1-3, SMO1-3, sterol 4-alpha methyl oxidase 1-3 |
-0.589 |
0.294 |
| 200 |
248471_at |
AT5G50840
|
unknown |
-0.579 |
0.091 |
| 201 |
261921_at |
AT1G65900
|
unknown |
-0.579 |
-0.102 |
| 202 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.576 |
0.047 |
| 203 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.575 |
-0.296 |
| 204 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
-0.575 |
-0.215 |
| 205 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
-0.565 |
-0.293 |
| 206 |
254633_at |
AT4G18640
|
MRH1, morphogenesis of root hair 1 |
-0.560 |
0.053 |
| 207 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
-0.559 |
-0.414 |
| 208 |
253294_at |
AT4G33740
|
unknown |
-0.557 |
0.143 |
| 209 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.555 |
0.003 |
| 210 |
248291_at |
AT5G53020
|
[Ribonuclease P protein subunit P38-related] |
-0.555 |
-0.011 |
| 211 |
263902_at |
AT2G36230
|
APG10, ALBINO AND PALE GREEN 10, HISN3 |
-0.550 |
-0.161 |
| 212 |
248178_at |
AT5G54370
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.549 |
-0.119 |
| 213 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.548 |
-0.009 |
| 214 |
247678_at |
AT5G59520
|
AtZIP2, ZIP2, ZRT/IRT-like protein 2 |
-0.545 |
0.163 |
| 215 |
260150_at |
AT1G52820
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.544 |
0.118 |
| 216 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.539 |
0.166 |
| 217 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.539 |
-0.402 |
| 218 |
247908_at |
AT5G57440
|
GPP2, GLYCEROL-3-PHOSPHATASE 2, GS1 |
-0.537 |
-0.156 |
| 219 |
251319_at |
AT3G61610
|
[Galactose mutarotase-like superfamily protein] |
-0.536 |
-0.149 |
| 220 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
-0.536 |
-0.207 |
| 221 |
263687_at |
AT1G26940
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.532 |
-0.139 |
| 222 |
257761_at |
AT3G23090
|
WDL3 |
-0.530 |
0.007 |
| 223 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.529 |
0.078 |
| 224 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
-0.529 |
-0.040 |
| 225 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.520 |
-0.037 |
| 226 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.514 |
-0.014 |
| 227 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.514 |
0.061 |
| 228 |
259080_at |
AT3G04910
|
ATWNK1, WNK1, with no lysine (K) kinase 1, ZIK4 |
-0.511 |
-0.082 |
| 229 |
255460_at |
AT4G02800
|
unknown |
-0.511 |
-0.449 |
| 230 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
-0.510 |
-0.174 |
| 231 |
265464_at |
AT2G37080
|
RIP2, ROP interactive partner 2 |
-0.509 |
0.017 |
| 232 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
-0.503 |
-0.070 |
| 233 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
-0.502 |
0.116 |
| 234 |
250689_at |
AT5G06610
|
unknown |
-0.501 |
-0.285 |
| 235 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
-0.501 |
0.126 |
| 236 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.500 |
0.058 |
| 237 |
267409_at |
AT2G34910
|
unknown |
-0.498 |
0.126 |
| 238 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
-0.497 |
0.078 |
| 239 |
244985_at |
ATCG00810
|
RPL22, ribosomal protein L22 |
-0.492 |
0.199 |
| 240 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.491 |
0.073 |
| 241 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
-0.490 |
-0.140 |
| 242 |
254412_at |
AT4G21430
|
B160 |
-0.489 |
-0.144 |
| 243 |
256602_at |
AT3G28310
|
unknown |
-0.488 |
0.012 |
| 244 |
258151_at |
AT3G18080
|
BGLU44, B-S glucosidase 44 |
-0.487 |
0.023 |
| 245 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
-0.485 |
0.049 |
| 246 |
248656_at |
AT5G48460
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.485 |
-0.319 |
| 247 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
-0.482 |
-0.073 |
| 248 |
245739_at |
AT1G44110
|
CYCA1;1, Cyclin A1;1 |
-0.481 |
-0.136 |
| 249 |
249886_at |
AT5G22320
|
[Leucine-rich repeat (LRR) family protein] |
-0.480 |
0.074 |
| 250 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
-0.475 |
0.204 |
| 251 |
263373_at |
AT2G20515
|
unknown |
-0.473 |
-0.032 |
| 252 |
258470_at |
AT3G06035
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.472 |
-0.051 |
| 253 |
248252_at |
AT5G53250
|
AGP22, arabinogalactan protein 22, ATAGP22, ARABINOGALACTAN PROTEIN 22 |
-0.472 |
-0.360 |
| 254 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
-0.468 |
-0.192 |
| 255 |
266226_at |
AT2G28740
|
HIS4, histone H4 |
-0.468 |
-0.143 |
| 256 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
-0.467 |
0.024 |
| 257 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
-0.466 |
-0.105 |
| 258 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.465 |
0.231 |
| 259 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
-0.462 |
-0.500 |
| 260 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
-0.457 |
0.005 |
| 261 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
-0.456 |
-0.348 |