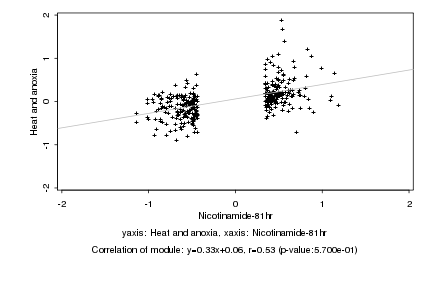
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Nicotinamide-81hr |
Log2 signal ratio
Heat and anoxia |
| 1 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
1.176 |
-0.085 |
| 2 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
1.132 |
0.660 |
| 3 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
1.099 |
0.137 |
| 4 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
1.090 |
0.028 |
| 5 |
248563_at |
AT5G49690
|
[UDP-Glycosyltransferase superfamily protein] |
0.990 |
0.777 |
| 6 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.899 |
-0.238 |
| 7 |
253859_at |
AT4G27657
|
unknown |
0.872 |
1.064 |
| 8 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.848 |
-0.159 |
| 9 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.833 |
0.052 |
| 10 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.824 |
1.205 |
| 11 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.814 |
0.582 |
| 12 |
260664_at |
AT1G19510
|
ATRL5, RAD-like 5, RL5, RAD-like 5, RSM4, RADIALIS-LIKE SANT/MYB 4 |
0.806 |
0.319 |
| 13 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.787 |
0.125 |
| 14 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.745 |
0.150 |
| 15 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
0.738 |
-0.155 |
| 16 |
259325_at |
AT3G05320
|
[O-fucosyltransferase family protein] |
0.734 |
0.276 |
| 17 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
0.730 |
0.160 |
| 18 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.716 |
0.226 |
| 19 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.699 |
-0.695 |
| 20 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.669 |
0.789 |
| 21 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.669 |
0.544 |
| 22 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.664 |
0.163 |
| 23 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.664 |
0.505 |
| 24 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.663 |
0.928 |
| 25 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
0.653 |
0.132 |
| 26 |
249454_at |
AT5G39520
|
unknown |
0.649 |
0.276 |
| 27 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.648 |
-0.142 |
| 28 |
254710_at |
AT4G18050
|
ABCB9, ATP-binding cassette B9, PGP9, P-glycoprotein 9 |
0.639 |
0.131 |
| 29 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
0.628 |
0.266 |
| 30 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
0.627 |
0.273 |
| 31 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
0.616 |
0.519 |
| 32 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.615 |
-0.055 |
| 33 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
0.615 |
0.201 |
| 34 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.608 |
0.255 |
| 35 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.605 |
0.212 |
| 36 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.602 |
-0.212 |
| 37 |
264375_at |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
0.601 |
0.431 |
| 38 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
0.599 |
0.160 |
| 39 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.579 |
0.091 |
| 40 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
0.574 |
-0.013 |
| 41 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.572 |
0.218 |
| 42 |
259150_at |
AT3G10320
|
[Glycosyltransferase family 61 protein] |
0.571 |
0.338 |
| 43 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.561 |
1.403 |
| 44 |
260933_at |
AT1G02470
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.554 |
-0.012 |
| 45 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
0.553 |
0.637 |
| 46 |
255430_at |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
0.551 |
0.620 |
| 47 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.548 |
0.489 |
| 48 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.545 |
-0.086 |
| 49 |
253182_at |
AT4G35190
|
LOG5, LONELY GUY 5 |
0.544 |
0.173 |
| 50 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.540 |
0.184 |
| 51 |
248777_at |
AT5G47920
|
unknown |
0.538 |
0.211 |
| 52 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
0.536 |
-0.203 |
| 53 |
259976_at |
AT1G76560
|
CP12-3, CP12 domain-containing protein 3 |
0.535 |
0.079 |
| 54 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.535 |
-0.199 |
| 55 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.532 |
1.670 |
| 56 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.529 |
0.448 |
| 57 |
245209_at |
AT5G12340
|
unknown |
0.527 |
0.730 |
| 58 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.527 |
1.897 |
| 59 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.526 |
0.159 |
| 60 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
0.524 |
0.169 |
| 61 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
0.524 |
0.203 |
| 62 |
258110_at |
AT3G14610
|
CYP72A7, cytochrome P450, family 72, subfamily A, polypeptide 7 |
0.521 |
-0.009 |
| 63 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.517 |
0.206 |
| 64 |
260337_at |
AT1G69310
|
ATWRKY57, WRKY57, WRKY DNA-binding protein 57 |
0.513 |
0.187 |
| 65 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.507 |
0.125 |
| 66 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
0.507 |
0.193 |
| 67 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
0.505 |
0.157 |
| 68 |
262228_at |
AT1G68690
|
AtPERK9, proline-rich extensin-like receptor kinase 9, PERK9, proline-rich extensin-like receptor kinase 9 |
0.503 |
0.346 |
| 69 |
247532_at |
AT5G61560
|
[U-box domain-containing protein kinase family protein] |
0.501 |
0.132 |
| 70 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.499 |
0.194 |
| 71 |
263702_at |
AT1G31240
|
[Bromodomain transcription factor] |
0.496 |
0.214 |
| 72 |
256818_at |
AT3G21420
|
LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1 |
0.496 |
0.156 |
| 73 |
245228_at |
AT3G29810
|
COBL2, COBRA-like protein 2 precursor |
0.495 |
0.508 |
| 74 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.494 |
0.511 |
| 75 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.493 |
1.094 |
| 76 |
259856_at |
AT1G68440
|
unknown |
0.490 |
0.196 |
| 77 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.489 |
0.681 |
| 78 |
248205_at |
AT5G54300
|
unknown |
0.487 |
0.809 |
| 79 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.486 |
0.375 |
| 80 |
256296_at |
AT1G69480
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.484 |
0.351 |
| 81 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
0.478 |
0.327 |
| 82 |
245420_at |
AT4G17410
|
[DWNN domain, a CCHC-type zinc finger] |
0.477 |
0.116 |
| 83 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.475 |
-0.036 |
| 84 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.475 |
0.135 |
| 85 |
249798_at |
AT5G23730
|
EFO2, EARLY FLOWERING BY OVEREXPRESSION 2, RUP2, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 2 |
0.471 |
0.052 |
| 86 |
247956_at |
AT5G56970
|
ATCKX3, CKX3, cytokinin oxidase 3 |
0.469 |
0.117 |
| 87 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
0.468 |
0.533 |
| 88 |
256574_at |
AT3G14780
|
unknown |
0.468 |
0.298 |
| 89 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
0.466 |
0.168 |
| 90 |
246418_at |
AT5G16960
|
[Zinc-binding dehydrogenase family protein] |
0.465 |
0.049 |
| 91 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
0.463 |
0.170 |
| 92 |
247212_at |
AT5G65040
|
unknown |
0.460 |
-0.125 |
| 93 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.460 |
0.022 |
| 94 |
263196_at |
AT1G36070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.460 |
0.144 |
| 95 |
257654_at |
AT3G13310
|
DJC66, DNA J protein C66 |
0.459 |
0.406 |
| 96 |
248207_at |
AT5G53970
|
TAT7, tyrosine aminotransferase 7 |
0.454 |
-0.018 |
| 97 |
260335_at |
AT1G74000
|
SS3, strictosidine synthase 3 |
0.453 |
-0.035 |
| 98 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
0.453 |
0.195 |
| 99 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.450 |
0.351 |
| 100 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.448 |
0.351 |
| 101 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
0.448 |
0.004 |
| 102 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
0.444 |
0.136 |
| 103 |
258170_at |
AT3G21600
|
[Senescence/dehydration-associated protein-related] |
0.441 |
0.021 |
| 104 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
0.435 |
-0.316 |
| 105 |
249190_at |
AT5G42750
|
BKI1, BRI1 kinase inhibitor 1 |
0.433 |
0.071 |
| 106 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.432 |
0.835 |
| 107 |
256767_at |
AT3G13680
|
[F-box and associated interaction domains-containing protein] |
0.429 |
0.246 |
| 108 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.427 |
0.221 |
| 109 |
255651_at |
AT4G00940
|
AtDOF4.1, DOF4.1, DNA binding with one finger 4.1, ITD1, INTERCELLULAR TRAFFICKING DOF 1 |
0.424 |
1.050 |
| 110 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
0.423 |
0.378 |
| 111 |
263380_at |
AT2G40200
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.422 |
0.043 |
| 112 |
255926_at |
AT1G22190
|
RAP2.4, related to AP2 4 |
0.422 |
0.414 |
| 113 |
263194_at |
AT1G36060
|
[Integrase-type DNA-binding superfamily protein] |
0.421 |
0.011 |
| 114 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.421 |
0.226 |
| 115 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.419 |
0.093 |
| 116 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.418 |
0.465 |
| 117 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.416 |
0.123 |
| 118 |
264288_at |
AT1G62045
|
unknown |
0.414 |
-0.174 |
| 119 |
261866_at |
AT1G50420
|
SCL-3, SCARECROW-LIKE 3, SCL3, scarecrow-like 3 |
0.409 |
-0.051 |
| 120 |
247848_at |
AT5G58120
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.406 |
0.128 |
| 121 |
246429_at |
AT5G17450
|
HIPP21, heavy metal associated isoprenylated plant protein 21 |
0.405 |
0.144 |
| 122 |
251621_at |
AT3G57700
|
[Protein kinase superfamily protein] |
0.403 |
-0.016 |
| 123 |
256321_at |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
0.403 |
0.006 |
| 124 |
261222_at |
AT1G20120
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.399 |
0.176 |
| 125 |
264452_at |
AT1G10270
|
GRP23, glutamine-rich protein 23 |
0.398 |
0.059 |
| 126 |
261161_at |
AT1G34420
|
[leucine-rich repeat transmembrane protein kinase family protein] |
0.398 |
0.070 |
| 127 |
255884_at |
AT1G20310
|
unknown |
0.396 |
0.188 |
| 128 |
257401_at |
AT1G23550
|
SRO2, similar to RCD one 2 |
0.395 |
0.921 |
| 129 |
263297_at |
AT2G15310
|
ARFB1A, ADP-ribosylation factor B1A, ATARFB1A, ADP-ribosylation factor B1A |
0.390 |
0.111 |
| 130 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
0.385 |
-0.132 |
| 131 |
260717_at |
AT1G48120
|
[hydrolases] |
0.385 |
0.122 |
| 132 |
264767_at |
AT1G61380
|
SD1-29, S-domain-1 29 |
0.384 |
-0.248 |
| 133 |
251727_at |
AT3G56290
|
unknown |
0.383 |
0.229 |
| 134 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
0.381 |
-0.134 |
| 135 |
252958_at |
AT4G38620
|
ATMYB4, myb domain protein 4, MYB4, myb domain protein 4 |
0.380 |
0.025 |
| 136 |
260327_at |
AT1G63840
|
[RING/U-box superfamily protein] |
0.380 |
0.325 |
| 137 |
247500_at |
AT5G61910
|
[DCD (Development and Cell Death) domain protein] |
0.379 |
0.020 |
| 138 |
247282_at |
AT5G64240
|
AtMC3, metacaspase 3, AtMCP1a, metacaspase 1a, MC3, metacaspase 3, MCP1a, metacaspase 1a |
0.375 |
0.176 |
| 139 |
265926_at |
AT2G18600
|
[Ubiquitin-conjugating enzyme family protein] |
0.374 |
0.086 |
| 140 |
248763_at |
AT5G47550
|
[Cystatin/monellin superfamily protein] |
0.373 |
-0.011 |
| 141 |
252822_at |
AT4G39955
|
[alpha/beta-Hydrolases superfamily protein] |
0.371 |
0.123 |
| 142 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
0.370 |
-0.114 |
| 143 |
261585_at |
AT1G01010
|
ANAC001, NAC domain containing protein 1, NAC001, NAC domain containing protein 1 |
0.368 |
0.086 |
| 144 |
247049_at |
AT5G66440
|
unknown |
0.367 |
-0.335 |
| 145 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
0.367 |
-0.079 |
| 146 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
0.366 |
-0.055 |
| 147 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.365 |
0.275 |
| 148 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
0.363 |
0.976 |
| 149 |
266775_at |
AT2G29065
|
[GRAS family transcription factor] |
0.360 |
0.429 |
| 150 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
0.357 |
0.323 |
| 151 |
259945_at |
AT1G71460
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.355 |
-0.198 |
| 152 |
252303_at |
AT3G49210
|
[O-acyltransferase (WSD1-like) family protein] |
0.355 |
-0.379 |
| 153 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
0.355 |
0.024 |
| 154 |
255881_at |
AT1G67070
|
DIN9, DARK INDUCIBLE 9, PMI2, PHOSPHOMANNOSE ISOMERASE 2 |
0.351 |
0.003 |
| 155 |
255578_at |
AT4G01450
|
UMAMIT30, Usually multiple acids move in and out Transporters 30 |
0.351 |
-0.135 |
| 156 |
246196_at |
AT4G37090
|
unknown |
0.348 |
0.198 |
| 157 |
246523_at |
AT5G15850
|
ATCOL1, BBX2, B-box domain protein 2, COL1, CONSTANS-like 1 |
0.346 |
0.429 |
| 158 |
260415_at |
AT1G69790
|
[Protein kinase superfamily protein] |
0.346 |
0.407 |
| 159 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
0.345 |
0.760 |
| 160 |
267066_at |
AT2G41040
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.344 |
0.015 |
| 161 |
247779_at |
AT5G58760
|
DDB2, damaged DNA binding 2 |
0.343 |
0.440 |
| 162 |
266269_at |
AT2G29480
|
ATGSTU2, glutathione S-transferase tau 2, GST20, GLUTATHIONE S-TRANSFERASE 20, GSTU2, glutathione S-transferase tau 2 |
0.343 |
0.867 |
| 163 |
266209_at |
AT2G27550
|
ATC, centroradialis |
0.343 |
-0.085 |
| 164 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.342 |
0.135 |
| 165 |
256861_at |
AT3G23920
|
AtBAM1, BAM1, beta-amylase 1, BMY7, BETA-AMYLASE 7, TR-BAMY |
0.341 |
0.582 |
| 166 |
260004_at |
AT1G67860
|
unknown |
0.340 |
0.190 |
| 167 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
0.340 |
0.194 |
| 168 |
260639_at |
AT1G53180
|
unknown |
0.340 |
0.324 |
| 169 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-1.151 |
-0.481 |
| 170 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-1.141 |
-0.267 |
| 171 |
250778_at |
AT5G05500
|
MOP10 |
-1.022 |
-0.369 |
| 172 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-1.017 |
-0.025 |
| 173 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-1.017 |
0.060 |
| 174 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-1.002 |
-0.412 |
| 175 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
-0.961 |
0.048 |
| 176 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
-0.955 |
-0.021 |
| 177 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.941 |
-0.782 |
| 178 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.941 |
0.171 |
| 179 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-0.927 |
-0.405 |
| 180 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
-0.915 |
-0.210 |
| 181 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.910 |
-0.626 |
| 182 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
-0.897 |
0.127 |
| 183 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.887 |
-0.182 |
| 184 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-0.887 |
0.158 |
| 185 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.875 |
0.141 |
| 186 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.869 |
-0.041 |
| 187 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.868 |
-0.448 |
| 188 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.865 |
-0.400 |
| 189 |
249545_at |
AT5G38030
|
[MATE efflux family protein] |
-0.858 |
-0.145 |
| 190 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.855 |
0.092 |
| 191 |
258080_at |
AT3G25930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.852 |
0.065 |
| 192 |
253720_at |
AT4G29270
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.849 |
-0.100 |
| 193 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.846 |
-0.483 |
| 194 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.841 |
0.212 |
| 195 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.823 |
-0.137 |
| 196 |
265050_at |
AT1G52070
|
[Mannose-binding lectin superfamily protein] |
-0.812 |
-0.232 |
| 197 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.799 |
-0.523 |
| 198 |
262598_at |
AT1G15260
|
unknown |
-0.799 |
-0.765 |
| 199 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.795 |
-0.151 |
| 200 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.788 |
-0.012 |
| 201 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
-0.787 |
0.115 |
| 202 |
250059_at |
AT5G17820
|
[Peroxidase superfamily protein] |
-0.782 |
-0.276 |
| 203 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-0.763 |
0.070 |
| 204 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.763 |
-0.104 |
| 205 |
267459_at |
AT2G33850
|
unknown |
-0.759 |
0.174 |
| 206 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.754 |
-0.671 |
| 207 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
-0.749 |
0.019 |
| 208 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.747 |
-0.113 |
| 209 |
258751_at |
AT3G05890
|
RCI2B, RARE-COLD-INDUCIBLE 2B |
-0.740 |
0.080 |
| 210 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.726 |
-0.362 |
| 211 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.719 |
0.119 |
| 212 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.702 |
-0.515 |
| 213 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-0.699 |
-0.145 |
| 214 |
247337_at |
AT5G63660
|
LCR74, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 74, PDF2.5 |
-0.697 |
-0.662 |
| 215 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
-0.697 |
0.391 |
| 216 |
266941_at |
AT2G18980
|
[Peroxidase superfamily protein] |
-0.690 |
-0.883 |
| 217 |
250157_at |
AT5G15180
|
[Peroxidase superfamily protein] |
-0.684 |
-0.392 |
| 218 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
-0.682 |
0.124 |
| 219 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.677 |
-0.242 |
| 220 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.676 |
-0.307 |
| 221 |
264987_at |
AT1G27030
|
unknown |
-0.673 |
0.150 |
| 222 |
266330_at |
AT2G01530
|
MLP329, MLP-like protein 329, ZCE2, (Zusammen-CA)-enhanced 2 |
-0.669 |
0.092 |
| 223 |
247678_at |
AT5G59520
|
AtZIP2, ZIP2, ZRT/IRT-like protein 2 |
-0.668 |
-0.062 |
| 224 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
-0.668 |
-0.219 |
| 225 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-0.661 |
-0.305 |
| 226 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
-0.658 |
-0.295 |
| 227 |
256878_at |
AT3G26460
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.650 |
0.149 |
| 228 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.639 |
-0.058 |
| 229 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
-0.636 |
-0.251 |
| 230 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.635 |
-0.178 |
| 231 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
-0.634 |
-0.599 |
| 232 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.634 |
-0.400 |
| 233 |
262236_at |
AT1G48330
|
unknown |
-0.626 |
-0.642 |
| 234 |
261921_at |
AT1G65900
|
unknown |
-0.626 |
-0.491 |
| 235 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
-0.624 |
0.100 |
| 236 |
250381_at |
AT5G11610
|
[Exostosin family protein] |
-0.622 |
0.157 |
| 237 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.620 |
-0.225 |
| 238 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
-0.612 |
-0.078 |
| 239 |
262700_at |
AT1G76020
|
[Thioredoxin superfamily protein] |
-0.610 |
0.118 |
| 240 |
249002_at |
AT5G44520
|
[NagB/RpiA/CoA transferase-like superfamily protein] |
-0.609 |
-0.193 |
| 241 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.607 |
0.025 |
| 242 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.607 |
-0.358 |
| 243 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.605 |
-0.091 |
| 244 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.605 |
0.064 |
| 245 |
265083_at |
AT1G03820
|
unknown |
-0.602 |
-0.555 |
| 246 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
-0.596 |
0.094 |
| 247 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-0.593 |
-0.489 |
| 248 |
254150_at |
AT4G24350
|
[Phosphorylase superfamily protein] |
-0.590 |
-0.065 |
| 249 |
253947_at |
AT4G26760
|
MAP65-2, microtubule-associated protein 65-2 |
-0.588 |
-0.288 |
| 250 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.580 |
0.141 |
| 251 |
262658_at |
AT1G14220
|
[Ribonuclease T2 family protein] |
-0.579 |
-0.345 |
| 252 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-0.578 |
0.333 |
| 253 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.577 |
-0.262 |
| 254 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.574 |
-0.009 |
| 255 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
-0.573 |
0.503 |
| 256 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-0.571 |
-0.327 |
| 257 |
253244_at |
AT4G34580
|
AtSFH1, COW1, CAN OF WORMS1, SRH1, SHORT ROOT HAIR 1 |
-0.569 |
-0.033 |
| 258 |
263902_at |
AT2G36230
|
APG10, ALBINO AND PALE GREEN 10, HISN3 |
-0.568 |
0.043 |
| 259 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
-0.568 |
-0.113 |
| 260 |
248178_at |
AT5G54370
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.563 |
-0.803 |
| 261 |
265248_at |
AT2G43010
|
AtPIF4, PIF4, phytochrome interacting factor 4, SRL2 |
-0.557 |
0.427 |
| 262 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.554 |
-0.223 |
| 263 |
254633_at |
AT4G18640
|
MRH1, morphogenesis of root hair 1 |
-0.554 |
-0.348 |
| 264 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.552 |
-0.011 |
| 265 |
261099_at |
AT1G62980
|
ATEXP18, ATEXPA18, expansin A18, ATHEXP ALPHA 1.25, EXP18, EXPANSIN 18, EXPA18, expansin A18 |
-0.549 |
-0.484 |
| 266 |
253421_at |
AT4G32340
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.548 |
0.004 |
| 267 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.547 |
-0.260 |
| 268 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
-0.543 |
-0.231 |
| 269 |
259091_at |
AT3G04890
|
[Uncharacterized conserved protein (DUF2358)] |
-0.539 |
-0.048 |
| 270 |
254573_at |
AT4G19420
|
[Pectinacetylesterase family protein] |
-0.536 |
-0.265 |
| 271 |
252077_at |
AT3G51720
|
unknown |
-0.536 |
-0.186 |
| 272 |
253613_at |
AT4G30320
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.532 |
-0.092 |
| 273 |
248252_at |
AT5G53250
|
AGP22, arabinogalactan protein 22, ATAGP22, ARABINOGALACTAN PROTEIN 22 |
-0.531 |
0.115 |
| 274 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
-0.523 |
-0.004 |
| 275 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
-0.521 |
-0.201 |
| 276 |
257761_at |
AT3G23090
|
WDL3 |
-0.518 |
0.199 |
| 277 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.515 |
-0.376 |
| 278 |
256585_at |
AT3G28760
|
unknown |
-0.514 |
0.002 |
| 279 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
-0.508 |
-0.176 |
| 280 |
261368_at |
AT1G53070
|
[Legume lectin family protein] |
-0.506 |
-0.267 |
| 281 |
247030_at |
AT5G67210
|
IRX15-L, IRX15-LIKE |
-0.505 |
-0.122 |
| 282 |
257924_at |
AT3G23190
|
[HR-like lesion-inducing protein-related] |
-0.502 |
-0.456 |
| 283 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.501 |
-0.027 |
| 284 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.499 |
-0.495 |
| 285 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.498 |
-0.514 |
| 286 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.497 |
0.025 |
| 287 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.495 |
-0.409 |
| 288 |
256021_at |
AT1G58270
|
ZW9 |
-0.495 |
-0.064 |
| 289 |
258376_at |
AT3G17680
|
[Kinase interacting (KIP1-like) family protein] |
-0.493 |
-0.066 |
| 290 |
247908_at |
AT5G57440
|
GPP2, GLYCEROL-3-PHOSPHATASE 2, GS1 |
-0.492 |
-0.155 |
| 291 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
-0.492 |
0.003 |
| 292 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.492 |
-0.462 |
| 293 |
257848_at |
AT3G13030
|
[hAT transposon superfamily protein] |
-0.492 |
-0.368 |
| 294 |
249970_at |
AT5G19100
|
[Eukaryotic aspartyl protease family protein] |
-0.491 |
0.322 |
| 295 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
-0.491 |
0.028 |
| 296 |
251996_at |
AT3G52840
|
BGAL2, beta-galactosidase 2 |
-0.491 |
0.071 |
| 297 |
251319_at |
AT3G61610
|
[Galactose mutarotase-like superfamily protein] |
-0.490 |
0.041 |
| 298 |
246487_at |
AT5G16030
|
unknown |
-0.489 |
-0.402 |
| 299 |
255876_at |
AT2G40480
|
unknown |
-0.489 |
-0.424 |
| 300 |
250043_at |
AT5G18430
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.488 |
-0.074 |
| 301 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
-0.487 |
0.196 |
| 302 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.486 |
0.163 |
| 303 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
-0.484 |
0.045 |
| 304 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
-0.484 |
-0.546 |
| 305 |
267191_at |
AT2G44110
|
ATMLO15, MILDEW RESISTANCE LOCUS O 15, MLO15, MILDEW RESISTANCE LOCUS O 15 |
-0.483 |
-0.398 |
| 306 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.483 |
-0.308 |
| 307 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
-0.482 |
0.046 |
| 308 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.482 |
-0.174 |
| 309 |
248656_at |
AT5G48460
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.481 |
-0.282 |
| 310 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
-0.480 |
0.134 |
| 311 |
253285_at |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
-0.477 |
0.014 |
| 312 |
263607_at |
AT2G16270
|
unknown |
-0.475 |
-0.048 |
| 313 |
265042_at |
AT1G04040
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.475 |
-0.065 |
| 314 |
257516_at |
AT1G69040
|
ACR4, ACT domain repeat 4 |
-0.475 |
-0.712 |
| 315 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.473 |
-0.136 |
| 316 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.469 |
0.117 |
| 317 |
254119_at |
AT4G24780
|
[Pectin lyase-like superfamily protein] |
-0.469 |
-0.606 |
| 318 |
265957_at |
AT2G37300
|
ABCI16, ATP-binding cassette I16 |
-0.461 |
-0.273 |
| 319 |
247094_at |
AT5G66280
|
GMD1, GDP-D-mannose 4,6-dehydratase 1 |
-0.460 |
-0.267 |
| 320 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
-0.459 |
-0.083 |
| 321 |
266396_at |
AT2G38790
|
unknown |
-0.458 |
0.629 |
| 322 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.457 |
-0.187 |
| 323 |
253353_at |
AT4G33730
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.455 |
-0.325 |
| 324 |
251142_at |
AT5G01015
|
unknown |
-0.455 |
0.378 |
| 325 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.452 |
-0.227 |
| 326 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.452 |
-0.258 |
| 327 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.452 |
-0.084 |
| 328 |
256441_at |
AT3G10940
|
LSF2, LIKE SEX4 2 |
-0.449 |
0.048 |
| 329 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
-0.447 |
0.122 |
| 330 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
-0.447 |
-0.285 |
| 331 |
263496_at |
AT2G42570
|
TBL39, TRICHOME BIREFRINGENCE-LIKE 39 |
-0.447 |
0.014 |
| 332 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
-0.446 |
-0.310 |
| 333 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
-0.446 |
-0.378 |
| 334 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.443 |
-0.697 |
| 335 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.443 |
-0.137 |