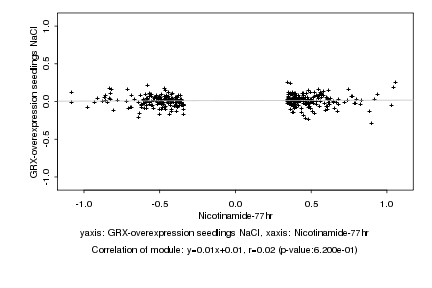
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Nicotinamide-77hr |
Log2 signal ratio
GRX-overexpression seedlings NaCl |
| 1 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
1.054 |
0.254 |
| 2 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
1.041 |
0.191 |
| 3 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
1.029 |
-0.047 |
| 4 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.934 |
0.096 |
| 5 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.914 |
0.030 |
| 6 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.895 |
-0.289 |
| 7 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.883 |
-0.126 |
| 8 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
0.831 |
0.026 |
| 9 |
253859_at |
AT4G27657
|
unknown |
0.824 |
-0.039 |
| 10 |
263918_at |
AT2G36590
|
ATPROT3, PROLINE TRANSPORTER 3, ProT3, proline transporter 3 |
0.799 |
0.036 |
| 11 |
260664_at |
AT1G19510
|
ATRL5, RAD-like 5, RL5, RAD-like 5, RSM4, RADIALIS-LIKE SANT/MYB 4 |
0.792 |
-0.014 |
| 12 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.783 |
-0.020 |
| 13 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
0.768 |
0.068 |
| 14 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.763 |
0.071 |
| 15 |
248563_at |
AT5G49690
|
[UDP-Glycosyltransferase superfamily protein] |
0.746 |
0.167 |
| 16 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.737 |
0.014 |
| 17 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.714 |
-0.007 |
| 18 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
0.680 |
0.033 |
| 19 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.674 |
-0.038 |
| 20 |
249747_at |
AT5G24600
|
unknown |
0.670 |
-0.129 |
| 21 |
260522_x_at |
AT2G41730
|
unknown |
0.666 |
-0.006 |
| 22 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.648 |
-0.080 |
| 23 |
263475_at |
AT2G31945
|
unknown |
0.645 |
-0.005 |
| 24 |
246584_at |
AT5G14730
|
unknown |
0.644 |
0.004 |
| 25 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.626 |
0.043 |
| 26 |
260744_at |
AT1G15010
|
unknown |
0.619 |
0.023 |
| 27 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.615 |
-0.030 |
| 28 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.612 |
0.035 |
| 29 |
266884_at |
AT2G44790
|
UCC2, uclacyanin 2 |
0.610 |
-0.004 |
| 30 |
256714_at |
AT2G34080
|
[Cysteine proteinases superfamily protein] |
0.609 |
0.149 |
| 31 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
0.607 |
0.053 |
| 32 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
0.607 |
-0.103 |
| 33 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
0.604 |
0.035 |
| 34 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.603 |
-0.058 |
| 35 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.596 |
0.062 |
| 36 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.595 |
-0.054 |
| 37 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.589 |
-0.108 |
| 38 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.585 |
-0.013 |
| 39 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.579 |
0.141 |
| 40 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
0.576 |
0.085 |
| 41 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
0.568 |
0.097 |
| 42 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.568 |
0.114 |
| 43 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
0.567 |
0.091 |
| 44 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.564 |
0.064 |
| 45 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.561 |
0.127 |
| 46 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.555 |
0.002 |
| 47 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
0.553 |
-0.062 |
| 48 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.552 |
0.103 |
| 49 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.550 |
0.082 |
| 50 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
0.547 |
0.166 |
| 51 |
255858_at |
AT1G67030
|
ZFP6, zinc finger protein 6 |
0.544 |
0.053 |
| 52 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.539 |
0.068 |
| 53 |
245382_at |
AT4G17800
|
AHL23, AT-hook motif nuclear localized protein 23 |
0.531 |
0.003 |
| 54 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
0.530 |
0.093 |
| 55 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.530 |
-0.032 |
| 56 |
252173_at |
AT3G50650
|
[GRAS family transcription factor] |
0.520 |
-0.149 |
| 57 |
258813_at |
AT3G04060
|
anac046, NAC domain containing protein 46, NAC046, NAC domain containing protein 46 |
0.519 |
0.121 |
| 58 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.516 |
0.071 |
| 59 |
267178_at |
AT2G37750
|
unknown |
0.515 |
0.054 |
| 60 |
257517_at |
AT3G16330
|
unknown |
0.513 |
0.010 |
| 61 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.510 |
-0.019 |
| 62 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.508 |
-0.125 |
| 63 |
258170_at |
AT3G21600
|
[Senescence/dehydration-associated protein-related] |
0.499 |
-0.009 |
| 64 |
260264_at |
AT1G68500
|
unknown |
0.499 |
0.039 |
| 65 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
0.495 |
-0.005 |
| 66 |
254410_at |
AT4G21410
|
CRK29, cysteine-rich RLK (RECEPTOR-like protein kinase) 29 |
0.494 |
-0.088 |
| 67 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.492 |
0.022 |
| 68 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
0.491 |
0.130 |
| 69 |
255926_at |
AT1G22190
|
RAP2.4, related to AP2 4 |
0.491 |
0.030 |
| 70 |
260337_at |
AT1G69310
|
ATWRKY57, WRKY57, WRKY DNA-binding protein 57 |
0.490 |
-0.089 |
| 71 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
0.486 |
-0.013 |
| 72 |
258752_at |
AT3G09520
|
ATEXO70H4, exocyst subunit exo70 family protein H4, EXO70H4, exocyst subunit exo70 family protein H4 |
0.485 |
-0.055 |
| 73 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
0.479 |
0.156 |
| 74 |
262801_at |
AT1G21010
|
unknown |
0.479 |
0.021 |
| 75 |
267307_at |
AT2G30210
|
LAC3, laccase 3 |
0.478 |
-0.028 |
| 76 |
266209_at |
AT2G27550
|
ATC, centroradialis |
0.477 |
0.036 |
| 77 |
247492_at |
AT5G61890
|
[Integrase-type DNA-binding superfamily protein] |
0.477 |
-0.228 |
| 78 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
0.477 |
-0.001 |
| 79 |
258110_at |
AT3G14610
|
CYP72A7, cytochrome P450, family 72, subfamily A, polypeptide 7 |
0.476 |
-0.066 |
| 80 |
250909_at |
AT5G03700
|
[D-mannose binding lectin protein with Apple-like carbohydrate-binding domain] |
0.475 |
-0.013 |
| 81 |
260261_at |
AT1G68450
|
PDE337, PIGMENT DEFECTIVE 337 |
0.474 |
0.057 |
| 82 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.469 |
-0.034 |
| 83 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.469 |
-0.029 |
| 84 |
251918_at |
AT3G54040
|
[PAR1 protein] |
0.467 |
-0.004 |
| 85 |
264685_at |
AT1G65610
|
ATGH9A2, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9A2, KOR2, KORRIGAN 2 |
0.463 |
0.113 |
| 86 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
0.462 |
0.057 |
| 87 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.459 |
-0.033 |
| 88 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.457 |
0.027 |
| 89 |
264082_at |
AT2G28570
|
unknown |
0.457 |
-0.215 |
| 90 |
261004_at |
AT1G26450
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.457 |
0.013 |
| 91 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
0.456 |
0.059 |
| 92 |
248225_at |
AT5G53740
|
unknown |
0.449 |
0.093 |
| 93 |
250777_at |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
0.447 |
0.011 |
| 94 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.445 |
0.111 |
| 95 |
263227_at |
AT1G30750
|
unknown |
0.445 |
-0.182 |
| 96 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.443 |
0.047 |
| 97 |
260335_at |
AT1G74000
|
SS3, strictosidine synthase 3 |
0.443 |
-0.021 |
| 98 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
0.440 |
0.050 |
| 99 |
249798_at |
AT5G23730
|
EFO2, EARLY FLOWERING BY OVEREXPRESSION 2, RUP2, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 2 |
0.440 |
0.111 |
| 100 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.436 |
-0.133 |
| 101 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
0.434 |
0.050 |
| 102 |
249258_at |
AT5G41650
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
0.430 |
-0.092 |
| 103 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
0.429 |
0.011 |
| 104 |
245420_at |
AT4G17410
|
[DWNN domain, a CCHC-type zinc finger] |
0.424 |
0.008 |
| 105 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.420 |
0.026 |
| 106 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
0.419 |
0.060 |
| 107 |
263797_at |
AT2G24570
|
ATWRKY17, WRKY17, WRKY DNA-binding protein 17 |
0.418 |
0.013 |
| 108 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.417 |
0.050 |
| 109 |
248496_at |
AT5G50790
|
AtSWEET10, SWEET10 |
0.416 |
0.086 |
| 110 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
0.415 |
0.046 |
| 111 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
0.414 |
0.058 |
| 112 |
267624_at |
AT2G39660
|
BIK1, botrytis-induced kinase1 |
0.414 |
-0.046 |
| 113 |
263931_at |
AT2G36220
|
unknown |
0.413 |
0.027 |
| 114 |
258323_at |
AT3G22750
|
[Protein kinase superfamily protein] |
0.408 |
0.020 |
| 115 |
245228_at |
AT3G29810
|
COBL2, COBRA-like protein 2 precursor |
0.407 |
-0.005 |
| 116 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.406 |
0.008 |
| 117 |
253830_at |
AT4G27652
|
unknown |
0.405 |
0.009 |
| 118 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
0.403 |
0.003 |
| 119 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.402 |
0.083 |
| 120 |
266280_at |
AT2G29260
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.402 |
0.052 |
| 121 |
255059_at |
AT4G09420
|
[Disease resistance protein (TIR-NBS class)] |
0.402 |
0.021 |
| 122 |
254208_at |
AT4G24175
|
unknown |
0.400 |
0.042 |
| 123 |
248763_at |
AT5G47550
|
[Cystatin/monellin superfamily protein] |
0.399 |
0.059 |
| 124 |
263194_at |
AT1G36060
|
[Integrase-type DNA-binding superfamily protein] |
0.399 |
-0.077 |
| 125 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.396 |
-0.030 |
| 126 |
245169_at |
AT2G33220
|
[GRIM-19 protein] |
0.395 |
0.019 |
| 127 |
246485_at |
AT5G16080
|
AtCXE17, carboxyesterase 17, CXE17, carboxyesterase 17 |
0.394 |
-0.029 |
| 128 |
265276_at |
AT2G28400
|
unknown |
0.393 |
0.027 |
| 129 |
247280_at |
AT5G64260
|
EXL2, EXORDIUM like 2 |
0.392 |
0.014 |
| 130 |
246984_at |
AT5G67310
|
CYP81G1, cytochrome P450, family 81, subfamily G, polypeptide 1 |
0.391 |
0.091 |
| 131 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.390 |
0.109 |
| 132 |
262844_at |
AT1G14890
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.388 |
-0.003 |
| 133 |
264941_at |
AT1G60680
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.388 |
0.007 |
| 134 |
264889_at |
AT1G23050
|
hydroxyproline-rich glycoprotein family protein |
0.387 |
-0.092 |
| 135 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
0.385 |
-0.017 |
| 136 |
265559_at |
AT2G05530
|
[Glycine-rich protein family] |
0.385 |
0.061 |
| 137 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
0.384 |
0.107 |
| 138 |
257946_at |
AT3G21710
|
unknown |
0.384 |
-0.058 |
| 139 |
250196_at |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
0.383 |
-0.057 |
| 140 |
264868_at |
AT1G24095
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.379 |
-0.033 |
| 141 |
249032_at |
AT5G44910
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.379 |
0.002 |
| 142 |
261481_at |
AT1G14260
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.379 |
-0.138 |
| 143 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
0.378 |
0.066 |
| 144 |
247049_at |
AT5G66440
|
unknown |
0.375 |
0.044 |
| 145 |
262038_at |
AT1G35580
|
A/N-InvG, alkaline/neutral invertase G, CINV1, cytosolic invertase 1 |
0.374 |
-0.044 |
| 146 |
266156_at |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
0.373 |
0.026 |
| 147 |
263297_at |
AT2G15310
|
ARFB1A, ADP-ribosylation factor B1A, ATARFB1A, ADP-ribosylation factor B1A |
0.372 |
0.097 |
| 148 |
252611_at |
AT3G45130
|
LAS1, lanosterol synthase 1 |
0.372 |
0.120 |
| 149 |
256377_at |
AT1G66650
|
[Protein with RING/U-box and TRAF-like domains] |
0.372 |
0.030 |
| 150 |
247052_at |
AT5G66700
|
ATHB53, ARABIDOPSIS THALIANA HOMEOBOX 53, HB-8, HOMEOBOX-8, HB53, homeobox 53 |
0.372 |
-0.144 |
| 151 |
252851_at |
AT4G40080
|
[ENTH/ANTH/VHS superfamily protein] |
0.371 |
-0.047 |
| 152 |
256321_at |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
0.368 |
-0.048 |
| 153 |
252214_at |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
0.367 |
0.096 |
| 154 |
249136_at |
AT5G43180
|
unknown |
0.367 |
-0.035 |
| 155 |
256433_at |
AT3G10985
|
ATWI-12, ARABIDOPSIS THALIANA WOUND-INDUCED PROTEIN 12, SAG20, senescence associated gene 20, WI12, WOUND-INDUCED PROTEIN 12 |
0.365 |
-0.025 |
| 156 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.365 |
0.033 |
| 157 |
266538_at |
AT2G35230
|
IKU1, HAIKU1 |
0.364 |
0.004 |
| 158 |
261756_at |
AT1G08315
|
[ARM repeat superfamily protein] |
0.363 |
0.035 |
| 159 |
250389_at |
AT5G11320
|
AtYUC4, YUC4, YUCCA4 |
0.363 |
0.244 |
| 160 |
260882_at |
AT1G29280
|
ATWRKY65, WRKY DNA-BINDING PROTEIN 65, WRKY65, WRKY DNA-binding protein 65 |
0.362 |
-0.030 |
| 161 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.360 |
0.107 |
| 162 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
0.359 |
0.012 |
| 163 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.358 |
-0.099 |
| 164 |
260023_at |
AT1G30040
|
ATGA2OX2, gibberellin 2-oxidase, GA2OX2, gibberellin 2-oxidase, GA2OX2, GIBBERELLIN 2-OXIDASE 2 |
0.357 |
0.102 |
| 165 |
257927_at |
AT3G23240
|
ATERF1, ETHYLENE RESPONSE FACTOR 1, ERF1, ethylene response factor 1 |
0.354 |
-0.037 |
| 166 |
247473_at |
AT5G62260
|
AHL6, AT-hook motif nuclear localized protein 6 |
0.354 |
-0.029 |
| 167 |
262947_at |
AT1G75750
|
GASA1, GAST1 protein homolog 1 |
0.350 |
0.012 |
| 168 |
264882_at |
AT1G61110
|
anac025, NAC domain containing protein 25, NAC025, NAC domain containing protein 25 |
0.350 |
0.075 |
| 169 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.349 |
0.000 |
| 170 |
250049_at |
AT5G17780
|
[alpha/beta-Hydrolases superfamily protein] |
0.349 |
0.101 |
| 171 |
245875_at |
AT1G26240
|
[Proline-rich extensin-like family protein] |
0.349 |
0.064 |
| 172 |
265943_at |
AT2G19570
|
AT-CDA1, CDA1, cytidine deaminase 1, DESZ |
0.347 |
0.003 |
| 173 |
267280_at |
AT2G19450
|
ABX45, AS11, ATDGAT, Arabidopsis thaliana acyl-CoA:diacylglycerol acyltransferase, AtDGAT1, DGAT1, acyl-CoA:diacylglycerol acyltransferase 1, RDS1, TAG1, TRIACYLGLYCEROL BIOSYNTHESIS DEFECT 1 |
0.347 |
0.030 |
| 174 |
245122_at |
AT2G47420
|
DIM1A, adenosine dimethyl transferase 1A |
0.346 |
-0.024 |
| 175 |
264645_at |
AT1G08940
|
[Phosphoglycerate mutase family protein] |
0.345 |
0.122 |
| 176 |
258795_at |
AT3G04570
|
AHL19, AT-hook motif nuclear-localized protein 19 |
0.344 |
0.051 |
| 177 |
261937_at |
AT1G22570
|
[Major facilitator superfamily protein] |
0.343 |
-0.016 |
| 178 |
264289_at |
AT1G61890
|
[MATE efflux family protein] |
0.343 |
0.004 |
| 179 |
258434_at |
AT3G16770
|
ATEBP, ethylene-responsive element binding protein, EBP, ethylene-responsive element binding protein, ERF72, ETHYLENE RESPONSE FACTOR 72, RAP2.3, RELATED TO AP2 3 |
0.342 |
0.037 |
| 180 |
258686_at |
AT3G07840
|
[Pectin lyase-like superfamily protein] |
0.342 |
0.262 |
| 181 |
258609_at |
AT3G02910
|
[AIG2-like (avirulence induced gene) family protein] |
0.342 |
0.014 |
| 182 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-1.087 |
0.126 |
| 183 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-1.085 |
-0.011 |
| 184 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-0.978 |
-0.075 |
| 185 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
-0.933 |
-0.005 |
| 186 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-0.916 |
0.045 |
| 187 |
252888_at |
AT4G39210
|
APL3 |
-0.880 |
0.009 |
| 188 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.866 |
0.065 |
| 189 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
-0.863 |
0.019 |
| 190 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.860 |
0.089 |
| 191 |
265063_at |
AT1G61500
|
[S-locus lectin protein kinase family protein] |
-0.851 |
-0.003 |
| 192 |
264698_at |
AT1G70200
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.833 |
0.045 |
| 193 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
-0.833 |
0.181 |
| 194 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.831 |
0.108 |
| 195 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
-0.828 |
0.028 |
| 196 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.819 |
0.160 |
| 197 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-0.810 |
-0.118 |
| 198 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.780 |
0.018 |
| 199 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
-0.721 |
0.012 |
| 200 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.713 |
0.161 |
| 201 |
249836_at |
AT5G23420
|
HMGB6, high-mobility group box 6 |
-0.710 |
-0.090 |
| 202 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.696 |
-0.073 |
| 203 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.689 |
0.088 |
| 204 |
244937_at |
ATCG01110
|
NDHH, NAD(P)H dehydrogenase subunit H |
-0.687 |
-0.068 |
| 205 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.671 |
0.034 |
| 206 |
250823_at |
AT5G05180
|
unknown |
-0.643 |
-0.004 |
| 207 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.640 |
-0.203 |
| 208 |
248150_at |
AT5G54670
|
ATK3, kinesin 3, KATC, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA C |
-0.639 |
-0.157 |
| 209 |
262137_at |
AT1G77920
|
TGA7, TGACG sequence-specific binding protein 7 |
-0.627 |
0.088 |
| 210 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
-0.626 |
-0.044 |
| 211 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.626 |
-0.031 |
| 212 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
-0.623 |
-0.073 |
| 213 |
258341_at |
AT3G22790
|
NET1A, Networked 1A |
-0.607 |
-0.024 |
| 214 |
251887_at |
AT3G54170
|
ATFIP37, FKBP12 interacting protein 37, FIP37, FKBP12 interacting protein 37 |
-0.607 |
0.036 |
| 215 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
-0.607 |
0.019 |
| 216 |
250233_at |
AT5G13460
|
IQD11, IQ-domain 11 |
-0.602 |
-0.084 |
| 217 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.601 |
-0.018 |
| 218 |
247425_at |
AT5G62550
|
unknown |
-0.598 |
-0.001 |
| 219 |
247942_at |
AT5G57120
|
unknown |
-0.597 |
0.104 |
| 220 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
-0.592 |
0.041 |
| 221 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.590 |
-0.049 |
| 222 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
-0.590 |
0.015 |
| 223 |
256878_at |
AT3G26460
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.589 |
-0.080 |
| 224 |
266589_at |
AT2G46250
|
[myosin heavy chain-related] |
-0.588 |
0.006 |
| 225 |
249863_at |
AT5G22950
|
VPS24.1 |
-0.585 |
0.049 |
| 226 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.584 |
0.002 |
| 227 |
259689_x_at |
AT1G63130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.584 |
0.003 |
| 228 |
265464_at |
AT2G37080
|
RIP2, ROP interactive partner 2 |
-0.583 |
0.034 |
| 229 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.581 |
0.046 |
| 230 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-0.581 |
0.215 |
| 231 |
256199_at |
AT1G58250
|
HPS4, hypersensitive to Pi starvation 4, SAB, SABRE |
-0.572 |
-0.008 |
| 232 |
262996_at |
AT1G54440
|
[Polynucleotidyl transferase, ribonuclease H fold protein with HRDC domain] |
-0.571 |
0.079 |
| 233 |
257848_at |
AT3G13030
|
[hAT transposon superfamily protein] |
-0.569 |
-0.033 |
| 234 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.569 |
0.107 |
| 235 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-0.563 |
0.095 |
| 236 |
252077_at |
AT3G51720
|
unknown |
-0.559 |
0.095 |
| 237 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.558 |
0.068 |
| 238 |
255460_at |
AT4G02800
|
unknown |
-0.557 |
0.005 |
| 239 |
258084_at |
AT3G26020
|
[Protein phosphatase 2A regulatory B subunit family protein] |
-0.556 |
-0.041 |
| 240 |
258080_at |
AT3G25930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.554 |
-0.045 |
| 241 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.547 |
0.023 |
| 242 |
265957_at |
AT2G37300
|
ABCI16, ATP-binding cassette I16 |
-0.543 |
-0.090 |
| 243 |
257761_at |
AT3G23090
|
WDL3 |
-0.539 |
0.053 |
| 244 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
-0.537 |
0.063 |
| 245 |
254412_at |
AT4G21430
|
B160 |
-0.530 |
0.061 |
| 246 |
245709_at |
AT5G04320
|
[Shugoshin C terminus] |
-0.523 |
0.007 |
| 247 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.522 |
0.092 |
| 248 |
248032_at |
AT5G55860
|
unknown |
-0.520 |
-0.007 |
| 249 |
247032_at |
AT5G67240
|
SDN3, small RNA degrading nuclease 3 |
-0.519 |
0.046 |
| 250 |
246231_at |
AT4G37080
|
unknown |
-0.516 |
-0.039 |
| 251 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
-0.514 |
0.031 |
| 252 |
258264_at |
AT3G15790
|
ATMBD11, MBD11, methyl-CPG-binding domain 11 |
-0.511 |
0.064 |
| 253 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.511 |
0.030 |
| 254 |
258744_at |
AT3G05830
|
unknown |
-0.510 |
0.029 |
| 255 |
257181_at |
AT3G13190
|
unknown |
-0.507 |
0.045 |
| 256 |
249545_at |
AT5G38030
|
[MATE efflux family protein] |
-0.506 |
-0.016 |
| 257 |
245460_at |
AT4G16990
|
RLM3, RESISTANCE TO LEPTOSPHAERIA MACULANS 3 |
-0.505 |
-0.105 |
| 258 |
262984_at |
AT1G54460
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.503 |
-0.005 |
| 259 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.502 |
0.072 |
| 260 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
-0.502 |
-0.159 |
| 261 |
266977_at |
AT2G39420
|
[alpha/beta-Hydrolases superfamily protein] |
-0.501 |
0.003 |
| 262 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.496 |
0.056 |
| 263 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
-0.495 |
-0.013 |
| 264 |
252916_at |
AT4G38950
|
[ATP binding microtubule motor family protein] |
-0.491 |
-0.031 |
| 265 |
263083_at |
AT2G27190
|
ATPAP1, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 1, ATPAP12, PURPLE ACID PHOSPHATASE 12, PAP1, PURPLE ACID PHOSPHATASE 1, PAP12, purple acid phosphatase 12 |
-0.491 |
0.027 |
| 266 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.489 |
0.106 |
| 267 |
258245_at |
AT3G29075
|
[glycine-rich protein] |
-0.489 |
0.031 |
| 268 |
247340_at |
AT5G63700
|
[zinc ion binding] |
-0.489 |
-0.094 |
| 269 |
261138_at |
AT1G19710
|
[UDP-Glycosyltransferase superfamily protein] |
-0.485 |
-0.071 |
| 270 |
248109_at |
AT5G55310
|
TOP1, TOPOISOMERASE 1, TOP1BETA, DNA topoisomerase 1 beta |
-0.485 |
-0.006 |
| 271 |
262538_at |
AT1G17140
|
ICR1, interactor of constitutive active rops 1, RIP1, ROP INTERACTIVE PARTNER 1 |
-0.484 |
-0.009 |
| 272 |
263544_at |
AT2G21590
|
APL4 |
-0.483 |
-0.013 |
| 273 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
-0.481 |
-0.005 |
| 274 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.477 |
-0.027 |
| 275 |
263545_at |
AT2G21560
|
unknown |
-0.474 |
0.077 |
| 276 |
258027_at |
AT3G19515
|
unknown |
-0.473 |
0.178 |
| 277 |
245861_at |
AT5G28300
|
AtGT2L, GT2L, GT-2Like protein |
-0.471 |
-0.074 |
| 278 |
259415_at |
AT1G02330
|
unknown |
-0.469 |
0.028 |
| 279 |
256052_at |
AT1G06960
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.466 |
-0.053 |
| 280 |
261421_at |
AT1G18840
|
IQD30, IQ-domain 30 |
-0.465 |
-0.093 |
| 281 |
265917_at |
AT2G15080
|
AtRLP19, receptor like protein 19, RLP19, receptor like protein 19 |
-0.465 |
0.050 |
| 282 |
254870_at |
AT4G11900
|
[S-locus lectin protein kinase family protein] |
-0.465 |
0.152 |
| 283 |
255008_at |
AT4G10060
|
[Beta-glucosidase, GBA2 type family protein] |
-0.464 |
0.037 |
| 284 |
255851_at |
AT1G67040
|
TRM22, TON1 Recruiting Motif 22 |
-0.464 |
0.052 |
| 285 |
266310_at |
AT2G26990
|
ATCSN2, COP12, CONSTITUTIVE PHOTOMORPHOGENIC 12, CSN2, FUS12, FUSCA 12 |
-0.461 |
0.058 |
| 286 |
249927_at |
AT5G19220
|
ADG2, ADP GLUCOSE PYROPHOSPHORYLASE 2, APL1, ADP glucose pyrophosphorylase large subunit 1 |
-0.458 |
0.040 |
| 287 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
-0.457 |
0.085 |
| 288 |
248106_at |
AT5G55100
|
[SWAP (Suppressor-of-White-APricot)/surp domain-containing protein] |
-0.456 |
0.034 |
| 289 |
246487_at |
AT5G16030
|
unknown |
-0.453 |
-0.009 |
| 290 |
245743_at |
AT1G51080
|
unknown |
-0.448 |
0.121 |
| 291 |
266117_at |
AT2G02170
|
[Remorin family protein] |
-0.447 |
0.028 |
| 292 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.446 |
0.017 |
| 293 |
250395_at |
AT5G10950
|
[Tudor/PWWP/MBT superfamily protein] |
-0.446 |
0.004 |
| 294 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.445 |
0.079 |
| 295 |
253337_at |
AT4G33470
|
ATHDA14, HDA14, histone deacetylase 14 |
-0.444 |
0.023 |
| 296 |
257295_at |
AT3G17420
|
GPK1, glyoxysomal protein kinase 1 |
-0.444 |
-0.022 |
| 297 |
259240_at |
AT3G11590
|
unknown |
-0.440 |
0.034 |
| 298 |
260697_at |
AT1G32530
|
[RING/U-box superfamily protein] |
-0.438 |
0.022 |
| 299 |
253720_at |
AT4G29270
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.437 |
-0.161 |
| 300 |
262598_at |
AT1G15260
|
unknown |
-0.437 |
0.027 |
| 301 |
249356_at |
AT5G40520
|
unknown |
-0.437 |
0.037 |
| 302 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
-0.432 |
-0.044 |
| 303 |
258300_at |
AT3G23340
|
ckl10, casein kinase I-like 10 |
-0.430 |
0.001 |
| 304 |
263569_at |
AT2G27170
|
SMC3, STRUCTURAL MAINTENANCE OF CHROMOSOMES 3, TTN7, TITAN7 |
-0.430 |
-0.023 |
| 305 |
259210_at |
AT3G09150
|
ATHY2, ARABIDOPSIS ELONGATED HYPOCOTYL 2, GUN3, GENOMES UNCOUPLED 3, HY2, ELONGATED HYPOCOTYL 2 |
-0.429 |
0.014 |
| 306 |
249002_at |
AT5G44520
|
[NagB/RpiA/CoA transferase-like superfamily protein] |
-0.427 |
0.095 |
| 307 |
261480_at |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-0.426 |
0.041 |
| 308 |
247197_at |
AT5G65240
|
[Leucine-rich repeat protein kinase family protein] |
-0.426 |
-0.128 |
| 309 |
249997_at |
AT5G18620
|
CHR17, chromatin remodeling factor17 |
-0.424 |
0.040 |
| 310 |
259207_at |
AT3G09050
|
unknown |
-0.423 |
0.027 |
| 311 |
263220_at |
AT1G30610
|
EMB2279, EMBRYO DEFECTIVE 2279, EMB88, EMBRYO DEFECTIVE 88 |
-0.423 |
-0.006 |
| 312 |
246415_at |
AT5G17160
|
unknown |
-0.423 |
-0.103 |
| 313 |
248378_at |
AT5G51840
|
unknown |
-0.419 |
0.050 |
| 314 |
265050_at |
AT1G52070
|
[Mannose-binding lectin superfamily protein] |
-0.419 |
0.112 |
| 315 |
247696_at |
AT5G59780
|
ATMYB59, MYB DOMAIN PROTEIN 59, ATMYB59-1, ATMYB59-2, ATMYB59-3, MYB59, myb domain protein 59 |
-0.418 |
0.018 |
| 316 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
-0.414 |
-0.000 |
| 317 |
251594_at |
AT3G57630
|
[exostosin family protein] |
-0.411 |
-0.011 |
| 318 |
254632_at |
AT4G18630
|
unknown |
-0.408 |
-0.053 |
| 319 |
255675_at |
AT4G00480
|
ATMYC1, myc1 |
-0.404 |
-0.058 |
| 320 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.404 |
-0.053 |
| 321 |
262275_at |
AT1G68710
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.401 |
0.000 |
| 322 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-0.401 |
-0.042 |
| 323 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
-0.397 |
0.033 |
| 324 |
254225_at |
AT4G23670
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.396 |
0.059 |
| 325 |
265305_at |
AT2G20340
|
AAS, aromatic aldehyde synthase, AtAAS |
-0.395 |
-0.046 |
| 326 |
262204_at |
AT2G01100
|
unknown |
-0.393 |
0.058 |
| 327 |
254638_at |
AT4G18740
|
[Rho termination factor] |
-0.393 |
-0.127 |
| 328 |
250689_at |
AT5G06610
|
unknown |
-0.393 |
0.016 |
| 329 |
246796_at |
AT5G26770
|
unknown |
-0.391 |
-0.011 |
| 330 |
260420_at |
AT1G69610
|
[FUNCTIONS IN: structural constituent of ribosome] |
-0.389 |
-0.072 |
| 331 |
261133_at |
AT1G19715
|
[Mannose-binding lectin superfamily protein] |
-0.388 |
0.033 |
| 332 |
246701_at |
AT5G28020
|
ATCYSD2, CYSTEINE SYNTHASE D2, CYSD2, cysteine synthase D2 |
-0.386 |
0.057 |
| 333 |
264800_at |
AT1G08800
|
MyoB1, myosin binding protein 1 |
-0.385 |
-0.053 |
| 334 |
248228_at |
AT5G53800
|
unknown |
-0.382 |
0.083 |
| 335 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
-0.379 |
0.011 |
| 336 |
257056_at |
AT3G15350
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.375 |
0.005 |
| 337 |
261117_at |
AT1G75310
|
AUL1, auxilin-like 1 |
-0.374 |
-0.049 |
| 338 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.371 |
0.005 |
| 339 |
263485_at |
AT2G29890
|
ATVLN1, VLN1, villin 1 |
-0.370 |
-0.020 |
| 340 |
255585_at |
AT4G01550
|
ANAC069, NAC domain containing protein 69, NAC069, NAC domain containing protein 69, NTM2, NAC with Transmembrane Motif 2 |
-0.370 |
-0.050 |
| 341 |
249404_at |
AT5G40160
|
EMB139, EMBRYO DEFECTIVE 139, EMB506, embryo defective 506 |
-0.369 |
-0.005 |
| 342 |
265330_at |
AT2G18440
|
GUT15, GENE WITH UNSTABLE TRANSCRIPT 15 |
-0.368 |
0.089 |
| 343 |
245845_at |
AT1G26150
|
AtPERK10, proline-rich extensin-like receptor kinase 10, PERK10, proline-rich extensin-like receptor kinase 10 |
-0.365 |
0.025 |
| 344 |
248094_at |
AT5G55220
|
[trigger factor type chaperone family protein] |
-0.363 |
0.033 |
| 345 |
261593_at |
AT1G33170
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.362 |
-0.008 |
| 346 |
267182_at |
AT2G23360
|
unknown |
-0.362 |
-0.009 |
| 347 |
257717_at |
AT3G18390
|
EMB1865, embryo defective 1865 |
-0.361 |
0.021 |
| 348 |
260974_at |
AT1G53440
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.360 |
0.022 |
| 349 |
249351_at |
AT5G40440
|
ATMKK3, mitogen-activated protein kinase kinase 3, MKK3, mitogen-activated protein kinase kinase 3 |
-0.359 |
-0.022 |
| 350 |
266489_at |
AT2G35190
|
ATNPSN11, NPSN11, novel plant snare 11, NSPN11 |
-0.358 |
-0.046 |
| 351 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.358 |
0.032 |
| 352 |
267538_at |
AT2G41870
|
[Remorin family protein] |
-0.357 |
0.025 |
| 353 |
249960_at |
AT5G18830
|
ATSPL7, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 7, SPL7, squamosa promoter binding protein-like 7 |
-0.355 |
-0.045 |
| 354 |
263700_at |
AT1G31150
|
[FUNCTIONS IN: sequence-specific DNA binding transcription factor activity] |
-0.354 |
-0.036 |
| 355 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.349 |
-0.098 |
| 356 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.346 |
-0.162 |
| 357 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
-0.346 |
-0.050 |
| 358 |
258355_at |
AT3G14330
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.346 |
-0.043 |
| 359 |
261471_at |
AT1G14460
|
[AAA-type ATPase family protein] |
-0.345 |
-0.043 |
| 360 |
253479_at |
AT4G32360
|
[Pyridine nucleotide-disulphide oxidoreductase family protein] |
-0.345 |
-0.028 |
| 361 |
249666_at |
AT5G35840
|
PHYC, phytochrome C |
-0.345 |
-0.031 |