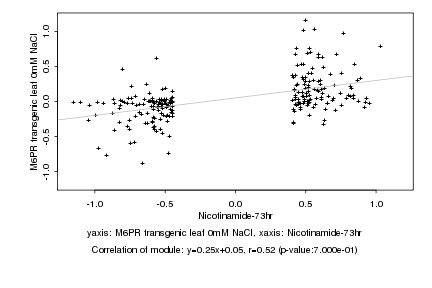
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Nicotinamide-73hr |
Log2 signal ratio
M6PR transgenic leaf 0mM NaCl |
| 1 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
1.030 |
0.794 |
| 2 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.948 |
-0.024 |
| 3 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
0.925 |
0.045 |
| 4 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
0.921 |
-0.006 |
| 5 |
261475_at |
AT1G14550
|
[Peroxidase superfamily protein] |
0.912 |
-0.083 |
| 6 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.885 |
0.339 |
| 7 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
0.868 |
0.008 |
| 8 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.862 |
0.301 |
| 9 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.840 |
0.542 |
| 10 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.837 |
0.044 |
| 11 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.833 |
0.091 |
| 12 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
0.821 |
0.195 |
| 13 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.818 |
0.227 |
| 14 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.814 |
0.077 |
| 15 |
249454_at |
AT5G39520
|
unknown |
0.797 |
0.092 |
| 16 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.787 |
0.051 |
| 17 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.763 |
0.977 |
| 18 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.752 |
0.402 |
| 19 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.751 |
-0.055 |
| 20 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
0.741 |
0.122 |
| 21 |
253322_at |
AT4G33980
|
unknown |
0.718 |
0.684 |
| 22 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.711 |
-0.123 |
| 23 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.699 |
0.243 |
| 24 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.696 |
0.054 |
| 25 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.687 |
0.015 |
| 26 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.673 |
0.396 |
| 27 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.659 |
0.083 |
| 28 |
264375_at |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
0.652 |
-0.018 |
| 29 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.651 |
-0.027 |
| 30 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.639 |
-0.100 |
| 31 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.632 |
0.192 |
| 32 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.629 |
-0.265 |
| 33 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.625 |
0.495 |
| 34 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
0.621 |
-0.323 |
| 35 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.618 |
0.635 |
| 36 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.612 |
0.122 |
| 37 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
0.611 |
0.036 |
| 38 |
259976_at |
AT1G76560
|
CP12-3, CP12 domain-containing protein 3 |
0.611 |
0.071 |
| 39 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
0.606 |
0.184 |
| 40 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.603 |
0.257 |
| 41 |
258813_at |
AT3G04060
|
anac046, NAC domain containing protein 46, NAC046, NAC domain containing protein 46 |
0.598 |
0.312 |
| 42 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.588 |
0.296 |
| 43 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
0.588 |
0.640 |
| 44 |
245209_at |
AT5G12340
|
unknown |
0.585 |
0.152 |
| 45 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
0.584 |
0.674 |
| 46 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
0.581 |
0.164 |
| 47 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
0.573 |
0.054 |
| 48 |
254014_at |
AT4G26120
|
[Ankyrin repeat family protein / BTB/POZ domain-containing protein] |
0.567 |
-0.042 |
| 49 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.557 |
0.182 |
| 50 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.556 |
1.041 |
| 51 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.554 |
-0.077 |
| 52 |
260522_x_at |
AT2G41730
|
unknown |
0.553 |
0.311 |
| 53 |
261937_at |
AT1G22570
|
[Major facilitator superfamily protein] |
0.544 |
0.479 |
| 54 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
0.540 |
0.404 |
| 55 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
0.528 |
0.040 |
| 56 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.528 |
0.711 |
| 57 |
267178_at |
AT2G37750
|
unknown |
0.526 |
0.090 |
| 58 |
263118_at |
AT1G03090
|
MCCA |
0.526 |
0.287 |
| 59 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.524 |
0.760 |
| 60 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.524 |
-0.008 |
| 61 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.520 |
-0.189 |
| 62 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.520 |
0.028 |
| 63 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.519 |
0.146 |
| 64 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.518 |
-0.022 |
| 65 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.517 |
0.234 |
| 66 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.516 |
0.211 |
| 67 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.515 |
0.409 |
| 68 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.510 |
0.693 |
| 69 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.510 |
0.003 |
| 70 |
248701_at |
AT5G48410
|
ATGLR1.3, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 1.3, GLR1.3, glutamate receptor 1.3 |
0.509 |
-0.045 |
| 71 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.505 |
0.124 |
| 72 |
244974_at |
ATCG00700
|
PSBN, photosystem II reaction center protein N |
0.499 |
0.081 |
| 73 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.496 |
1.171 |
| 74 |
265481_at |
AT2G15960
|
unknown |
0.495 |
0.294 |
| 75 |
249032_at |
AT5G44910
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.492 |
0.351 |
| 76 |
259511_at |
AT1G12520
|
ATCCS, copper chaperone for SOD1, CCS, copper chaperone for SOD1 |
0.492 |
0.197 |
| 77 |
257950_at |
AT3G21780
|
UGT71B6, UDP-glucosyl transferase 71B6 |
0.488 |
0.035 |
| 78 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
0.487 |
0.012 |
| 79 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.484 |
0.252 |
| 80 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.480 |
-0.025 |
| 81 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.480 |
0.077 |
| 82 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.478 |
0.325 |
| 83 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
0.478 |
1.017 |
| 84 |
254408_at |
AT4G21390
|
B120 |
0.477 |
0.530 |
| 85 |
261558_at |
AT1G01770
|
unknown |
0.473 |
0.082 |
| 86 |
259945_at |
AT1G71460
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.472 |
0.140 |
| 87 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
0.471 |
0.346 |
| 88 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.468 |
0.532 |
| 89 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.460 |
-0.074 |
| 90 |
267226_at |
AT2G44010
|
unknown |
0.457 |
-0.023 |
| 91 |
258170_at |
AT3G21600
|
[Senescence/dehydration-associated protein-related] |
0.456 |
-0.100 |
| 92 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.452 |
-0.027 |
| 93 |
257333_at |
ATMG01360
|
COX1, cytochrome oxidase |
0.447 |
-0.032 |
| 94 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
0.447 |
0.036 |
| 95 |
264543_at |
AT1G55790
|
[FUNCTIONS IN: metal ion binding] |
0.445 |
-0.007 |
| 96 |
266884_at |
AT2G44790
|
UCC2, uclacyanin 2 |
0.445 |
0.143 |
| 97 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
0.441 |
0.516 |
| 98 |
264645_at |
AT1G08940
|
[Phosphoglycerate mutase family protein] |
0.439 |
0.249 |
| 99 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
0.432 |
0.771 |
| 100 |
248298_at |
AT5G53110
|
[RING/U-box superfamily protein] |
0.431 |
-0.052 |
| 101 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
0.430 |
0.236 |
| 102 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
0.424 |
0.678 |
| 103 |
247282_at |
AT5G64240
|
AtMC3, metacaspase 3, AtMCP1a, metacaspase 1a, MC3, metacaspase 3, MCP1a, metacaspase 1a |
0.424 |
0.089 |
| 104 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.424 |
-0.036 |
| 105 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.421 |
0.042 |
| 106 |
248896_at |
AT5G46350
|
ATWRKY8, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 8, WRKY8, WRKY DNA-binding protein 8 |
0.421 |
0.061 |
| 107 |
261934_at |
AT1G22400
|
ATUGT85A1, ARABIDOPSIS THALIANA UDP-GLUCOSYL TRANSFERASE 85A1, UGT85A1 |
0.420 |
0.377 |
| 108 |
264746_at |
AT1G62300
|
ATWRKY6, WRKY6 |
0.419 |
0.168 |
| 109 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.418 |
-0.140 |
| 110 |
248934_at |
AT5G46080
|
[Protein kinase superfamily protein] |
0.415 |
0.181 |
| 111 |
262228_at |
AT1G68690
|
AtPERK9, proline-rich extensin-like receptor kinase 9, PERK9, proline-rich extensin-like receptor kinase 9 |
0.411 |
0.348 |
| 112 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.410 |
-0.292 |
| 113 |
263157_at |
AT1G54100
|
ALDH7B4, aldehyde dehydrogenase 7B4 |
0.409 |
-0.111 |
| 114 |
259293_at |
AT3G11580
|
[AP2/B3-like transcriptional factor family protein] |
0.407 |
-0.313 |
| 115 |
266231_at |
AT2G02220
|
ATPSKR1, PHYTOSULFOKIN RECEPTOR 1, PSKR1, phytosulfokin receptor 1 |
0.405 |
0.381 |
| 116 |
256818_at |
AT3G21420
|
LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1 |
0.404 |
0.022 |
| 117 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-1.156 |
-0.009 |
| 118 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-1.107 |
-0.013 |
| 119 |
252888_at |
AT4G39210
|
APL3 |
-1.052 |
-0.271 |
| 120 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-1.041 |
-0.051 |
| 121 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.998 |
-0.198 |
| 122 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-0.987 |
-0.005 |
| 123 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.979 |
-0.667 |
| 124 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.942 |
-0.024 |
| 125 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.923 |
-0.763 |
| 126 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.878 |
-0.162 |
| 127 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
-0.869 |
0.029 |
| 128 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.865 |
-0.410 |
| 129 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.861 |
-0.026 |
| 130 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.830 |
-0.088 |
| 131 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
-0.826 |
-0.287 |
| 132 |
259399_at |
AT1G17710
|
AtPEPC1, Arabidopsis thaliana phosphoethanolamine/phosphocholine phosphatase 1, PEPC1, phosphoethanolamine/phosphocholine phosphatase 1 |
-0.821 |
-0.044 |
| 133 |
250778_at |
AT5G05500
|
MOP10 |
-0.819 |
0.023 |
| 134 |
249004_at |
AT5G44570
|
unknown |
-0.809 |
0.009 |
| 135 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.808 |
0.471 |
| 136 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
-0.792 |
-0.008 |
| 137 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
-0.772 |
-0.021 |
| 138 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.771 |
-0.350 |
| 139 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.764 |
0.054 |
| 140 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.760 |
-0.261 |
| 141 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.755 |
-0.389 |
| 142 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
-0.753 |
-0.590 |
| 143 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.746 |
0.047 |
| 144 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
-0.742 |
0.226 |
| 145 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-0.736 |
-0.022 |
| 146 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.719 |
-0.579 |
| 147 |
253720_at |
AT4G29270
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.715 |
0.075 |
| 148 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.712 |
-0.208 |
| 149 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.706 |
-0.052 |
| 150 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-0.689 |
-0.146 |
| 151 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.688 |
-0.036 |
| 152 |
262598_at |
AT1G15260
|
unknown |
-0.674 |
-0.184 |
| 153 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-0.663 |
-0.876 |
| 154 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.657 |
-0.029 |
| 155 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.652 |
0.029 |
| 156 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.643 |
-0.309 |
| 157 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
-0.636 |
0.249 |
| 158 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.631 |
-0.159 |
| 159 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.629 |
-0.306 |
| 160 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.622 |
0.014 |
| 161 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.618 |
0.010 |
| 162 |
257421_at |
AT1G12030
|
unknown |
-0.616 |
0.118 |
| 163 |
263902_at |
AT2G36230
|
APG10, ALBINO AND PALE GREEN 10, HISN3 |
-0.605 |
0.009 |
| 164 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.598 |
-0.067 |
| 165 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
-0.596 |
-0.299 |
| 166 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.594 |
0.051 |
| 167 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
-0.594 |
0.004 |
| 168 |
255460_at |
AT4G02800
|
unknown |
-0.592 |
0.019 |
| 169 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
-0.592 |
-0.281 |
| 170 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.589 |
-0.005 |
| 171 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.587 |
-0.364 |
| 172 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.587 |
-0.228 |
| 173 |
253947_at |
AT4G26760
|
MAP65-2, microtubule-associated protein 65-2 |
-0.586 |
-0.113 |
| 174 |
261684_at |
AT1G47400
|
unknown |
-0.586 |
-0.063 |
| 175 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
-0.582 |
-0.350 |
| 176 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.581 |
-0.235 |
| 177 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.579 |
-0.388 |
| 178 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
-0.578 |
-0.239 |
| 179 |
258080_at |
AT3G25930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.578 |
-0.014 |
| 180 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
-0.568 |
-0.421 |
| 181 |
263083_at |
AT2G27190
|
ATPAP1, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 1, ATPAP12, PURPLE ACID PHOSPHATASE 12, PAP1, PURPLE ACID PHOSPHATASE 1, PAP12, purple acid phosphatase 12 |
-0.568 |
0.010 |
| 182 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.564 |
0.626 |
| 183 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
-0.560 |
-0.072 |
| 184 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.554 |
-0.065 |
| 185 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.554 |
-0.114 |
| 186 |
256441_at |
AT3G10940
|
LSF2, LIKE SEX4 2 |
-0.548 |
-0.040 |
| 187 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.547 |
-0.008 |
| 188 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-0.542 |
0.038 |
| 189 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.540 |
-0.391 |
| 190 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.538 |
-0.248 |
| 191 |
247337_at |
AT5G63660
|
LCR74, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 74, PDF2.5 |
-0.536 |
-0.015 |
| 192 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
-0.532 |
-0.171 |
| 193 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
-0.531 |
-0.066 |
| 194 |
259091_at |
AT3G04890
|
[Uncharacterized conserved protein (DUF2358)] |
-0.530 |
0.022 |
| 195 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.529 |
0.031 |
| 196 |
261368_at |
AT1G53070
|
[Legume lectin family protein] |
-0.524 |
-0.089 |
| 197 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.524 |
0.181 |
| 198 |
262651_at |
AT1G14100
|
FUT8, fucosyltransferase 8 |
-0.522 |
-0.033 |
| 199 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
-0.519 |
-0.452 |
| 200 |
258341_at |
AT3G22790
|
NET1A, Networked 1A |
-0.519 |
-0.033 |
| 201 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.513 |
-0.191 |
| 202 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.508 |
0.007 |
| 203 |
253244_at |
AT4G34580
|
AtSFH1, COW1, CAN OF WORMS1, SRH1, SHORT ROOT HAIR 1 |
-0.507 |
0.020 |
| 204 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
-0.505 |
0.042 |
| 205 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
-0.504 |
0.194 |
| 206 |
255876_at |
AT2G40480
|
unknown |
-0.502 |
-0.019 |
| 207 |
258264_at |
AT3G15790
|
ATMBD11, MBD11, methyl-CPG-binding domain 11 |
-0.495 |
-0.205 |
| 208 |
266413_at |
AT2G38740
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.493 |
-0.076 |
| 209 |
256585_at |
AT3G28760
|
unknown |
-0.492 |
-0.034 |
| 210 |
255958_at |
AT1G22150
|
SULTR1;3, sulfate transporter 1;3 |
-0.492 |
0.039 |
| 211 |
259276_at |
AT3G01190
|
[Peroxidase superfamily protein] |
-0.489 |
-0.066 |
| 212 |
245348_at |
AT4G17770
|
ATTPS5, trehalose phosphatase/synthase 5, TPS5, TREHALOSE -6-PHOSPHATASE SYNTHASE S5, TPS5, trehalose phosphatase/synthase 5 |
-0.488 |
-0.279 |
| 213 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.487 |
-0.190 |
| 214 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.482 |
-0.731 |
| 215 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.477 |
-0.052 |
| 216 |
252365_at |
AT3G48350
|
CEP3, cysteine endopeptidase 3 |
-0.471 |
-0.009 |
| 217 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.471 |
-0.210 |
| 218 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-0.469 |
-0.486 |
| 219 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
-0.466 |
-0.020 |
| 220 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
-0.465 |
-0.102 |
| 221 |
263687_at |
AT1G26940
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.463 |
0.018 |
| 222 |
249588_at |
AT5G37790
|
[Protein kinase superfamily protein] |
-0.461 |
-0.143 |
| 223 |
247908_at |
AT5G57440
|
GPP2, GLYCEROL-3-PHOSPHATASE 2, GS1 |
-0.460 |
0.057 |
| 224 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.453 |
-0.068 |
| 225 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.453 |
-0.121 |
| 226 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.452 |
-0.160 |
| 227 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.452 |
0.060 |
| 228 |
254633_at |
AT4G18640
|
MRH1, morphogenesis of root hair 1 |
-0.450 |
0.006 |
| 229 |
249002_at |
AT5G44520
|
[NagB/RpiA/CoA transferase-like superfamily protein] |
-0.450 |
-0.013 |
| 230 |
252089_at |
AT3G52110
|
unknown |
-0.450 |
0.151 |
| 231 |
263782_at |
AT2G46380
|
unknown |
-0.450 |
-0.207 |