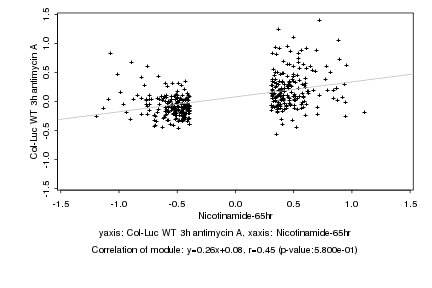
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Nicotinamide-65hr |
Log2 signal ratio
Col-Luc WT 3h antimycin A |
| 1 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
1.105 |
-0.181 |
| 2 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.953 |
0.624 |
| 3 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.944 |
0.000 |
| 4 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
0.937 |
-0.243 |
| 5 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.936 |
0.296 |
| 6 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.913 |
0.071 |
| 7 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
0.885 |
0.735 |
| 8 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
0.883 |
1.053 |
| 9 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.876 |
0.025 |
| 10 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.862 |
0.232 |
| 11 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
0.862 |
0.238 |
| 12 |
248563_at |
AT5G49690
|
[UDP-Glycosyltransferase superfamily protein] |
0.842 |
0.193 |
| 13 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.837 |
0.069 |
| 14 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.811 |
0.510 |
| 15 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.785 |
0.207 |
| 16 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
0.779 |
0.615 |
| 17 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
0.771 |
0.382 |
| 18 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.715 |
1.414 |
| 19 |
264375_at |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
0.715 |
0.108 |
| 20 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.701 |
-0.094 |
| 21 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.697 |
-0.213 |
| 22 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.691 |
0.888 |
| 23 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.685 |
0.518 |
| 24 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
0.662 |
0.201 |
| 25 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.654 |
0.540 |
| 26 |
247532_at |
AT5G61560
|
[U-box domain-containing protein kinase family protein] |
0.638 |
0.616 |
| 27 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.624 |
0.190 |
| 28 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.622 |
0.148 |
| 29 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
0.611 |
0.190 |
| 30 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.605 |
0.918 |
| 31 |
267493_at |
AT2G30400
|
ATOFP2, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 2, OFP2, ovate family protein 2 |
0.603 |
0.584 |
| 32 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
0.602 |
-0.037 |
| 33 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
0.599 |
0.248 |
| 34 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.599 |
-0.241 |
| 35 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.599 |
0.410 |
| 36 |
259976_at |
AT1G76560
|
CP12-3, CP12 domain-containing protein 3 |
0.595 |
0.317 |
| 37 |
266269_at |
AT2G29480
|
ATGSTU2, glutathione S-transferase tau 2, GST20, GLUTATHIONE S-TRANSFERASE 20, GSTU2, glutathione S-transferase tau 2 |
0.590 |
0.277 |
| 38 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.586 |
0.013 |
| 39 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
0.581 |
-0.100 |
| 40 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.581 |
0.649 |
| 41 |
246418_at |
AT5G16960
|
[Zinc-binding dehydrogenase family protein] |
0.581 |
0.103 |
| 42 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
0.580 |
0.072 |
| 43 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.575 |
-0.013 |
| 44 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.572 |
0.310 |
| 45 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.572 |
0.148 |
| 46 |
252539_at |
AT3G45730
|
unknown |
0.565 |
0.897 |
| 47 |
259511_at |
AT1G12520
|
ATCCS, copper chaperone for SOD1, CCS, copper chaperone for SOD1 |
0.565 |
-0.115 |
| 48 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
0.563 |
-0.038 |
| 49 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
0.547 |
0.375 |
| 50 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.546 |
0.586 |
| 51 |
256170_at |
AT1G51790
|
[Leucine-rich repeat protein kinase family protein] |
0.544 |
0.102 |
| 52 |
246370_at |
AT1G51920
|
unknown |
0.540 |
0.747 |
| 53 |
245840_at |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
0.540 |
0.686 |
| 54 |
262228_at |
AT1G68690
|
AtPERK9, proline-rich extensin-like receptor kinase 9, PERK9, proline-rich extensin-like receptor kinase 9 |
0.538 |
0.842 |
| 55 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.536 |
0.231 |
| 56 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
0.531 |
-0.131 |
| 57 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
0.526 |
0.064 |
| 58 |
254410_at |
AT4G21410
|
CRK29, cysteine-rich RLK (RECEPTOR-like protein kinase) 29 |
0.525 |
0.464 |
| 59 |
263593_at |
AT2G01860
|
EMB975, EMBRYO DEFECTIVE 975 |
0.520 |
-0.445 |
| 60 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
0.515 |
0.329 |
| 61 |
258468_at |
AT3G06070
|
unknown |
0.512 |
0.025 |
| 62 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
0.508 |
0.095 |
| 63 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
0.507 |
0.031 |
| 64 |
247052_at |
AT5G66700
|
ATHB53, ARABIDOPSIS THALIANA HOMEOBOX 53, HB-8, HOMEOBOX-8, HB53, homeobox 53 |
0.505 |
0.122 |
| 65 |
250971_at |
AT5G02810
|
APRR7, PRR7, pseudo-response regulator 7 |
0.501 |
0.050 |
| 66 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
0.499 |
-0.053 |
| 67 |
248777_at |
AT5G47920
|
unknown |
0.499 |
-0.110 |
| 68 |
253827_at |
AT4G28085
|
unknown |
0.498 |
1.116 |
| 69 |
264685_at |
AT1G65610
|
ATGH9A2, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9A2, KOR2, KORRIGAN 2 |
0.497 |
0.479 |
| 70 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
0.495 |
0.446 |
| 71 |
250335_at |
AT5G11650
|
[alpha/beta-Hydrolases superfamily protein] |
0.494 |
0.359 |
| 72 |
249765_at |
AT5G24030
|
SLAH3, SLAC1 homologue 3 |
0.493 |
0.443 |
| 73 |
256098_at |
AT1G13700
|
PGL1, 6-phosphogluconolactonase 1 |
0.492 |
0.099 |
| 74 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.491 |
0.616 |
| 75 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
0.487 |
-0.326 |
| 76 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
0.486 |
0.381 |
| 77 |
246790_at |
AT5G27610
|
ALY1, ALWAYS EARLY 1, ATALY1, ARABIDOPSIS THALIANA ALWAYS EARLY 1 |
0.475 |
0.268 |
| 78 |
263118_at |
AT1G03090
|
MCCA |
0.475 |
0.159 |
| 79 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.471 |
-0.041 |
| 80 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.469 |
0.873 |
| 81 |
266165_at |
AT2G28190
|
CSD2, copper/zinc superoxide dismutase 2, CZSOD2, COPPER/ZINC SUPEROXIDE DISMUTASE 2 |
0.469 |
-0.057 |
| 82 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.467 |
0.279 |
| 83 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
0.464 |
0.066 |
| 84 |
250323_at |
AT5G12880
|
[proline-rich family protein] |
0.464 |
0.654 |
| 85 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.463 |
-0.021 |
| 86 |
265086_at |
AT1G03770
|
ATRING1B, ARABIDOPSIS THALIANA RING 1B, RING1B, RING 1B |
0.462 |
0.330 |
| 87 |
258170_at |
AT3G21600
|
[Senescence/dehydration-associated protein-related] |
0.461 |
0.030 |
| 88 |
265999_at |
AT2G24100
|
ASG1, ALTERED SEED GERMINATION 1 |
0.459 |
0.349 |
| 89 |
246978_at |
AT5G24910
|
CYP714A1, cytochrome P450, family 714, subfamily A, polypeptide 1, ELA1, EUI-like p450 A1 |
0.453 |
0.080 |
| 90 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
0.450 |
0.271 |
| 91 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.450 |
0.234 |
| 92 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.449 |
0.286 |
| 93 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
0.449 |
0.307 |
| 94 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.448 |
0.240 |
| 95 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.445 |
0.444 |
| 96 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.442 |
0.276 |
| 97 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.441 |
0.651 |
| 98 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.441 |
0.960 |
| 99 |
264082_at |
AT2G28570
|
unknown |
0.439 |
0.105 |
| 100 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
0.431 |
-0.064 |
| 101 |
258323_at |
AT3G22750
|
[Protein kinase superfamily protein] |
0.431 |
-0.114 |
| 102 |
248496_at |
AT5G50790
|
AtSWEET10, SWEET10 |
0.430 |
-0.078 |
| 103 |
261558_at |
AT1G01770
|
unknown |
0.419 |
-0.129 |
| 104 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.418 |
0.232 |
| 105 |
264809_at |
AT1G08830
|
CSD1, copper/zinc superoxide dismutase 1 |
0.415 |
-0.028 |
| 106 |
246943_at |
AT5G25440
|
[Protein kinase superfamily protein] |
0.414 |
0.190 |
| 107 |
247282_at |
AT5G64240
|
AtMC3, metacaspase 3, AtMCP1a, metacaspase 1a, MC3, metacaspase 3, MCP1a, metacaspase 1a |
0.413 |
0.296 |
| 108 |
248934_at |
AT5G46080
|
[Protein kinase superfamily protein] |
0.412 |
0.695 |
| 109 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
0.411 |
0.019 |
| 110 |
264767_at |
AT1G61380
|
SD1-29, S-domain-1 29 |
0.411 |
0.116 |
| 111 |
264867_at |
AT1G24150
|
ATFH4, FORMIN HOMOLOGUE 4, FH4, formin homologue 4 |
0.410 |
0.490 |
| 112 |
260517_at |
AT1G51420
|
ATSPP1, SUCROSE-PHOSPHATASE 1, SPP1, sucrose-phosphatase 1 |
0.410 |
0.044 |
| 113 |
252822_at |
AT4G39955
|
[alpha/beta-Hydrolases superfamily protein] |
0.410 |
0.110 |
| 114 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
0.407 |
0.055 |
| 115 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.404 |
-0.167 |
| 116 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.403 |
-0.386 |
| 117 |
250803_at |
AT5G04980
|
[DNAse I-like superfamily protein] |
0.402 |
0.207 |
| 118 |
264756_at |
AT1G61370
|
[S-locus lectin protein kinase family protein] |
0.402 |
0.332 |
| 119 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
0.401 |
0.499 |
| 120 |
263157_at |
AT1G54100
|
ALDH7B4, aldehyde dehydrogenase 7B4 |
0.400 |
0.058 |
| 121 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.399 |
0.356 |
| 122 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.395 |
0.213 |
| 123 |
257479_at |
AT1G16150
|
WAKL4, wall associated kinase-like 4 |
0.395 |
0.187 |
| 124 |
265494_at |
AT2G15680
|
AtCML30, CML30, calmodulin-like 30 |
0.394 |
0.041 |
| 125 |
263196_at |
AT1G36070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.394 |
0.031 |
| 126 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
0.387 |
0.234 |
| 127 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.387 |
-0.304 |
| 128 |
258110_at |
AT3G14610
|
CYP72A7, cytochrome P450, family 72, subfamily A, polypeptide 7 |
0.386 |
0.093 |
| 129 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
0.385 |
0.479 |
| 130 |
261222_at |
AT1G20120
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.383 |
-0.082 |
| 131 |
249136_at |
AT5G43180
|
unknown |
0.381 |
-0.045 |
| 132 |
263570_at |
AT2G27150
|
AAO3, abscisic aldehyde oxidase 3, AOdelta, Aldehyde oxidase delta, At-AO3, Arabidopsis thaliana aldehyde oxidase 3, AtAAO3 |
0.381 |
0.145 |
| 133 |
247903_at |
AT5G57340
|
unknown |
0.380 |
-0.115 |
| 134 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
0.380 |
-0.112 |
| 135 |
264645_at |
AT1G08940
|
[Phosphoglycerate mutase family protein] |
0.378 |
0.346 |
| 136 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.378 |
0.925 |
| 137 |
248207_at |
AT5G53970
|
TAT7, tyrosine aminotransferase 7 |
0.375 |
0.119 |
| 138 |
261585_at |
AT1G01010
|
ANAC001, NAC domain containing protein 1, NAC001, NAC domain containing protein 1 |
0.374 |
0.127 |
| 139 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
0.372 |
-0.059 |
| 140 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
0.370 |
0.207 |
| 141 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.368 |
0.309 |
| 142 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.367 |
0.197 |
| 143 |
263702_at |
AT1G31240
|
[Bromodomain transcription factor] |
0.366 |
-0.002 |
| 144 |
251647_at |
AT3G57770
|
[Protein kinase superfamily protein] |
0.366 |
-0.066 |
| 145 |
256818_at |
AT3G21420
|
LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1 |
0.365 |
-0.106 |
| 146 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
0.363 |
0.117 |
| 147 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.362 |
1.246 |
| 148 |
259711_at |
AT1G77570
|
AtREN1, REN1, restricted to nucleolus 1 |
0.358 |
-0.123 |
| 149 |
247212_at |
AT5G65040
|
unknown |
0.357 |
-0.062 |
| 150 |
258188_at |
AT3G17800
|
unknown |
0.357 |
0.097 |
| 151 |
249798_at |
AT5G23730
|
EFO2, EARLY FLOWERING BY OVEREXPRESSION 2, RUP2, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 2 |
0.356 |
0.100 |
| 152 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.356 |
-0.161 |
| 153 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
0.355 |
-0.129 |
| 154 |
246144_at |
AT5G20110
|
[Dynein light chain type 1 family protein] |
0.355 |
-0.188 |
| 155 |
259856_at |
AT1G68440
|
unknown |
0.353 |
0.506 |
| 156 |
256981_at |
AT3G13380
|
BRL3, BRI1-like 3 |
0.351 |
0.316 |
| 157 |
259579_at |
AT1G28010
|
ABCB14, ATP-binding cassette B14, ATABCB14, Arabidopsis thaliana ATP-binding cassette B14, MDR12, multi-drug resistance 12, PGP14, P-glycoprotein 14 |
0.349 |
-0.565 |
| 158 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
0.349 |
0.071 |
| 159 |
247005_at |
AT5G67520
|
adenosine-5'-phosphosulfate (APS) kinase 4 |
0.347 |
-0.074 |
| 160 |
267140_at |
AT2G38250
|
[Homeodomain-like superfamily protein] |
0.345 |
0.820 |
| 161 |
254496_at |
AT4G20070
|
AAH, allantoate amidohydrolase, ATAAH, allantoate amidohydrolase |
0.344 |
0.272 |
| 162 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.343 |
0.946 |
| 163 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
0.343 |
0.248 |
| 164 |
249677_at |
AT5G35970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.342 |
0.029 |
| 165 |
261544_at |
AT1G63540
|
[hydroxyproline-rich glycoprotein family protein] |
0.341 |
0.067 |
| 166 |
264746_at |
AT1G62300
|
ATWRKY6, WRKY6 |
0.341 |
0.515 |
| 167 |
248763_at |
AT5G47550
|
[Cystatin/monellin superfamily protein] |
0.339 |
0.009 |
| 168 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.336 |
0.383 |
| 169 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
0.336 |
0.074 |
| 170 |
245056_at |
AT2G26480
|
UGT76D1, UDP-glucosyl transferase 76D1 |
0.333 |
0.142 |
| 171 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
0.332 |
-0.102 |
| 172 |
258262_at |
AT3G15770
|
unknown |
0.332 |
0.333 |
| 173 |
264261_at |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
0.328 |
0.154 |
| 174 |
254014_at |
AT4G26120
|
[Ankyrin repeat family protein / BTB/POZ domain-containing protein] |
0.328 |
0.462 |
| 175 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
0.326 |
0.125 |
| 176 |
261674_at |
AT1G18270
|
[ketose-bisphosphate aldolase class-II family protein] |
0.326 |
0.004 |
| 177 |
260956_at |
AT1G06040
|
BBX24, B-box domain protein 24, STO, SALT TOLERANCE |
0.321 |
-0.074 |
| 178 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.319 |
0.556 |
| 179 |
246999_at |
AT5G67440
|
MEL2, MAB4/ENP/NPY1-LIKE 2, NPY3, NAKED PINS IN YUC MUTANTS 3 |
0.319 |
0.127 |
| 180 |
260337_at |
AT1G69310
|
ATWRKY57, WRKY57, WRKY DNA-binding protein 57 |
0.318 |
0.058 |
| 181 |
253113_at |
AT4G35985
|
[Senescence/dehydration-associated protein-related] |
0.318 |
0.391 |
| 182 |
265470_at |
AT2G37150
|
[RING/U-box superfamily protein] |
0.316 |
0.192 |
| 183 |
267261_at |
AT2G23120
|
[Late embryogenesis abundant protein, group 6] |
0.316 |
0.209 |
| 184 |
261934_at |
AT1G22400
|
ATUGT85A1, ARABIDOPSIS THALIANA UDP-GLUCOSYL TRANSFERASE 85A1, UGT85A1 |
0.316 |
0.352 |
| 185 |
267467_at |
AT2G30580
|
BMI1A, DRIP2, DREB2A-interacting protein 2 |
0.315 |
0.066 |
| 186 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
0.315 |
0.088 |
| 187 |
247956_at |
AT5G56970
|
ATCKX3, CKX3, cytokinin oxidase 3 |
0.314 |
0.112 |
| 188 |
254681_at |
AT4G18140
|
SSP4b, SCP1-like small phosphatase 4b |
0.314 |
0.281 |
| 189 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.312 |
0.830 |
| 190 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
0.310 |
0.146 |
| 191 |
262456_at |
AT1G11260
|
ATSTP1, SUGAR TRANSPORTER 1, STP1, sugar transporter 1 |
0.310 |
0.149 |
| 192 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
0.310 |
-0.091 |
| 193 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
0.309 |
-0.055 |
| 194 |
255602_at |
AT4G01026
|
PYL7, PYR1-like 7, RCAR2, regulatory components of ABA receptor 2 |
0.309 |
-0.138 |
| 195 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
0.309 |
0.291 |
| 196 |
244977_at |
ATCG00730
|
PETD, photosynthetic electron transfer D |
0.309 |
0.152 |
| 197 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
0.308 |
-0.053 |
| 198 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-1.199 |
-0.249 |
| 199 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-1.140 |
-0.111 |
| 200 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-1.095 |
0.049 |
| 201 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-1.077 |
0.842 |
| 202 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
-1.020 |
0.476 |
| 203 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.995 |
0.156 |
| 204 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.969 |
-0.041 |
| 205 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.938 |
-0.189 |
| 206 |
252888_at |
AT4G39210
|
APL3 |
-0.905 |
-0.296 |
| 207 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.898 |
0.682 |
| 208 |
249004_at |
AT5G44570
|
unknown |
-0.876 |
0.046 |
| 209 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.847 |
0.121 |
| 210 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
-0.815 |
-0.211 |
| 211 |
250778_at |
AT5G05500
|
MOP10 |
-0.815 |
0.067 |
| 212 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.808 |
0.417 |
| 213 |
246001_at |
AT5G20790
|
unknown |
-0.782 |
0.293 |
| 214 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.768 |
0.035 |
| 215 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
-0.765 |
-0.040 |
| 216 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.764 |
0.034 |
| 217 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
-0.761 |
-0.077 |
| 218 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
-0.759 |
0.621 |
| 219 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.756 |
-0.221 |
| 220 |
262651_at |
AT1G14100
|
FUT8, fucosyltransferase 8 |
-0.745 |
0.036 |
| 221 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.745 |
0.114 |
| 222 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.740 |
-0.038 |
| 223 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.739 |
-0.138 |
| 224 |
267317_at |
AT2G34700
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.721 |
0.050 |
| 225 |
265083_at |
AT1G03820
|
unknown |
-0.700 |
-0.325 |
| 226 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.699 |
-0.225 |
| 227 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.699 |
-0.418 |
| 228 |
261921_at |
AT1G65900
|
unknown |
-0.699 |
-0.226 |
| 229 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.694 |
-0.402 |
| 230 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.690 |
-0.258 |
| 231 |
262598_at |
AT1G15260
|
unknown |
-0.679 |
-0.329 |
| 232 |
261368_at |
AT1G53070
|
[Legume lectin family protein] |
-0.672 |
-0.188 |
| 233 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.671 |
0.059 |
| 234 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
-0.666 |
0.441 |
| 235 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.663 |
-0.129 |
| 236 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.655 |
-0.046 |
| 237 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-0.655 |
0.037 |
| 238 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.640 |
0.102 |
| 239 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.632 |
-0.438 |
| 240 |
267555_at |
AT2G32765
|
ATSUMO5, SUM5, SUMO 5, SUMO5, small ubiquitinrelated modifier 5 |
-0.628 |
0.025 |
| 241 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.623 |
0.061 |
| 242 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.619 |
-0.283 |
| 243 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
-0.617 |
-0.275 |
| 244 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.615 |
0.091 |
| 245 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.613 |
-0.267 |
| 246 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
-0.606 |
0.315 |
| 247 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-0.605 |
0.007 |
| 248 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
-0.605 |
-0.164 |
| 249 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.604 |
-0.096 |
| 250 |
264998_at |
AT1G67330
|
unknown |
-0.602 |
-0.230 |
| 251 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.600 |
0.086 |
| 252 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.599 |
-0.213 |
| 253 |
265305_at |
AT2G20340
|
AAS, aromatic aldehyde synthase, AtAAS |
-0.595 |
-0.127 |
| 254 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
-0.594 |
0.109 |
| 255 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.589 |
-0.018 |
| 256 |
253940_at |
AT4G26950
|
unknown |
-0.588 |
0.273 |
| 257 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.586 |
-0.312 |
| 258 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.584 |
-0.163 |
| 259 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.581 |
-0.321 |
| 260 |
263902_at |
AT2G36230
|
APG10, ALBINO AND PALE GREEN 10, HISN3 |
-0.579 |
-0.160 |
| 261 |
255958_at |
AT1G22150
|
SULTR1;3, sulfate transporter 1;3 |
-0.579 |
0.013 |
| 262 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
-0.578 |
-0.080 |
| 263 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
-0.575 |
-0.089 |
| 264 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.575 |
-0.185 |
| 265 |
250059_at |
AT5G17820
|
[Peroxidase superfamily protein] |
-0.568 |
-0.007 |
| 266 |
255876_at |
AT2G40480
|
unknown |
-0.563 |
-0.395 |
| 267 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
-0.562 |
0.013 |
| 268 |
266089_at |
AT2G38010
|
[Neutral/alkaline non-lysosomal ceramidase] |
-0.556 |
-0.101 |
| 269 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
-0.554 |
-0.147 |
| 270 |
262700_at |
AT1G76020
|
[Thioredoxin superfamily protein] |
-0.553 |
0.093 |
| 271 |
263628_at |
AT2G04780
|
FLA7, FASCICLIN-like arabinoogalactan 7 |
-0.551 |
-0.053 |
| 272 |
264987_at |
AT1G27030
|
unknown |
-0.550 |
0.042 |
| 273 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.548 |
-0.046 |
| 274 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
-0.548 |
-0.111 |
| 275 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.546 |
0.314 |
| 276 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.540 |
-0.410 |
| 277 |
249167_at |
AT5G42860
|
unknown |
-0.540 |
-0.200 |
| 278 |
253244_at |
AT4G34580
|
AtSFH1, COW1, CAN OF WORMS1, SRH1, SHORT ROOT HAIR 1 |
-0.538 |
-0.115 |
| 279 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
-0.536 |
-0.207 |
| 280 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.530 |
0.089 |
| 281 |
252916_at |
AT4G38950
|
[ATP binding microtubule motor family protein] |
-0.529 |
-0.129 |
| 282 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.527 |
-0.204 |
| 283 |
266941_at |
AT2G18980
|
[Peroxidase superfamily protein] |
-0.526 |
-0.069 |
| 284 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.523 |
0.121 |
| 285 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.523 |
-0.118 |
| 286 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
-0.523 |
0.049 |
| 287 |
266418_at |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
-0.522 |
0.035 |
| 288 |
265957_at |
AT2G37300
|
ABCI16, ATP-binding cassette I16 |
-0.522 |
-0.173 |
| 289 |
263640_at |
AT2G25270
|
unknown |
-0.521 |
-0.226 |
| 290 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.521 |
-0.298 |
| 291 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.520 |
0.049 |
| 292 |
265656_at |
AT2G13820
|
AtXYP2, XYP2, xylogen protein 2 |
-0.516 |
-0.168 |
| 293 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
-0.515 |
-0.148 |
| 294 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.514 |
-0.199 |
| 295 |
248596_at |
AT5G49330
|
ATMYB111, ARABIDOPSIS MYB DOMAIN PROTEIN 111, MYB111, myb domain protein 111, PFG3, PRODUCTION OF FLAVONOL GLYCOSIDES 3 |
-0.514 |
-0.224 |
| 296 |
265740_at |
AT2G01150
|
RHA2B, RING-H2 finger protein 2B |
-0.513 |
0.136 |
| 297 |
247425_at |
AT5G62550
|
unknown |
-0.511 |
-0.134 |
| 298 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.511 |
-0.283 |
| 299 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.509 |
-0.154 |
| 300 |
251143_at |
AT5G01220
|
SQD2, sulfoquinovosyldiacylglycerol 2 |
-0.507 |
-0.028 |
| 301 |
251142_at |
AT5G01015
|
unknown |
-0.506 |
0.113 |
| 302 |
248074_at |
AT5G55730
|
FLA1, FASCICLIN-like arabinogalactan 1 |
-0.504 |
-0.071 |
| 303 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.503 |
0.190 |
| 304 |
259091_at |
AT3G04890
|
[Uncharacterized conserved protein (DUF2358)] |
-0.501 |
-0.075 |
| 305 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
-0.500 |
0.001 |
| 306 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
-0.499 |
0.057 |
| 307 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.498 |
0.081 |
| 308 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.498 |
-0.007 |
| 309 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.497 |
-0.132 |
| 310 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.497 |
-0.338 |
| 311 |
249002_at |
AT5G44520
|
[NagB/RpiA/CoA transferase-like superfamily protein] |
-0.496 |
-0.103 |
| 312 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-0.496 |
-0.340 |
| 313 |
254290_at |
AT4G23000
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.496 |
-0.007 |
| 314 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.494 |
-0.460 |
| 315 |
263083_at |
AT2G27190
|
ATPAP1, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 1, ATPAP12, PURPLE ACID PHOSPHATASE 12, PAP1, PURPLE ACID PHOSPHATASE 1, PAP12, purple acid phosphatase 12 |
-0.494 |
-0.083 |
| 316 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
-0.491 |
0.317 |
| 317 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
-0.489 |
-0.153 |
| 318 |
261806_at |
AT1G30510
|
ATRFNR2, root FNR 2, RFNR2, root FNR 2 |
-0.486 |
0.020 |
| 319 |
260041_at |
AT1G68780
|
[RNI-like superfamily protein] |
-0.483 |
-0.229 |
| 320 |
245348_at |
AT4G17770
|
ATTPS5, trehalose phosphatase/synthase 5, TPS5, TREHALOSE -6-PHOSPHATASE SYNTHASE S5, TPS5, trehalose phosphatase/synthase 5 |
-0.482 |
-0.313 |
| 321 |
253337_at |
AT4G33470
|
ATHDA14, HDA14, histone deacetylase 14 |
-0.480 |
-0.180 |
| 322 |
246652_at |
AT5G35190
|
EXT13, extensin 13 |
-0.474 |
0.065 |
| 323 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.470 |
-0.324 |
| 324 |
249392_at |
AT5G40150
|
[Peroxidase superfamily protein] |
-0.470 |
-0.234 |
| 325 |
251982_at |
AT3G53190
|
[Pectin lyase-like superfamily protein] |
-0.469 |
-0.156 |
| 326 |
251493_at |
AT3G59300
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.467 |
-0.038 |
| 327 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
-0.465 |
0.282 |
| 328 |
253729_at |
AT4G29360
|
[O-Glycosyl hydrolases family 17 protein] |
-0.465 |
-0.095 |
| 329 |
252168_at |
AT3G50440
|
ATMES10, ARABIDOPSIS THALIANA METHYL ESTERASE 10, MES10, methyl esterase 10 |
-0.464 |
-0.195 |
| 330 |
254333_at |
AT4G22753
|
ATSMO1-3, SMO1-3, sterol 4-alpha methyl oxidase 1-3 |
-0.464 |
0.038 |
| 331 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-0.462 |
-0.320 |
| 332 |
251395_at |
AT2G45470
|
AGP8, ARABINOGALACTAN PROTEIN 8, FLA8, FASCICLIN-like arabinogalactan protein 8 |
-0.459 |
-0.164 |
| 333 |
259014_at |
AT3G07320
|
[O-Glycosyl hydrolases family 17 protein] |
-0.457 |
-0.032 |
| 334 |
260034_at |
AT1G68810
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.456 |
0.104 |
| 335 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.455 |
-0.249 |
| 336 |
246159_at |
AT5G20935
|
CRR42, CHLORORESPIRATORY REDUCTION 42 |
-0.455 |
-0.331 |
| 337 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.452 |
-0.110 |
| 338 |
246797_at |
AT5G26790
|
unknown |
-0.452 |
-0.072 |
| 339 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.449 |
-0.306 |
| 340 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.448 |
0.023 |
| 341 |
263915_at |
AT2G36430
|
unknown |
-0.447 |
-0.125 |
| 342 |
254119_at |
AT4G24780
|
[Pectin lyase-like superfamily protein] |
-0.447 |
-0.290 |
| 343 |
246983_at |
AT5G67200
|
[Leucine-rich repeat protein kinase family protein] |
-0.447 |
-0.317 |
| 344 |
258633_at |
AT3G07990
|
SCPL27, serine carboxypeptidase-like 27 |
-0.447 |
-0.188 |
| 345 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
-0.444 |
0.208 |
| 346 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.444 |
-0.208 |
| 347 |
255924_at |
AT1G22170
|
[Phosphoglycerate mutase family protein] |
-0.440 |
-0.099 |
| 348 |
265277_at |
AT2G28410
|
unknown |
-0.440 |
-0.304 |
| 349 |
264839_at |
AT1G03630
|
POR C, protochlorophyllide oxidoreductase C, PORC |
-0.439 |
-0.241 |
| 350 |
256400_at |
AT3G06140
|
LUL4, LOG2-LIKE UBIQUITIN LIGASE4 |
-0.438 |
-0.044 |
| 351 |
247077_at |
AT5G66420
|
unknown |
-0.438 |
-0.081 |
| 352 |
249046_at |
AT5G44400
|
[FAD-binding Berberine family protein] |
-0.436 |
0.028 |
| 353 |
252149_at |
AT3G51290
|
APSR1, Altered Phosphate Starvation Response 1 |
-0.436 |
-0.192 |
| 354 |
263544_at |
AT2G21590
|
APL4 |
-0.436 |
-0.274 |
| 355 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
-0.434 |
0.361 |
| 356 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
-0.433 |
0.046 |
| 357 |
267590_at |
AT2G39700
|
ATEXP4, ATEXPA4, expansin A4, ATHEXP ALPHA 1.6, EXPA4, expansin A4 |
-0.433 |
-0.088 |
| 358 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.427 |
0.055 |
| 359 |
258082_at |
AT3G25905
|
CLE27, CLAVATA3/ESR-RELATED 27 |
-0.427 |
-0.113 |
| 360 |
253753_at |
AT4G29030
|
[Putative membrane lipoprotein] |
-0.427 |
-0.048 |
| 361 |
247541_at |
AT5G61660
|
[glycine-rich protein] |
-0.426 |
-0.092 |
| 362 |
247101_at |
AT5G66530
|
[Galactose mutarotase-like superfamily protein] |
-0.426 |
-0.060 |
| 363 |
254794_at |
AT4G12970
|
EPFL9, STOMAGEN, STOMAGEN |
-0.426 |
-0.192 |
| 364 |
251843_x_at |
AT3G54590
|
ATHRGP1, hydroxyproline-rich glycoprotein, HRGP1, hydroxyproline-rich glycoprotein |
-0.420 |
0.143 |
| 365 |
247239_at |
AT5G64640
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.419 |
-0.136 |
| 366 |
256052_at |
AT1G06960
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.418 |
0.049 |
| 367 |
257474_at |
AT1G80850
|
[DNA glycosylase superfamily protein] |
-0.417 |
-0.084 |
| 368 |
255488_at |
AT4G02630
|
[Protein kinase superfamily protein] |
-0.415 |
-0.142 |
| 369 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.414 |
-0.341 |
| 370 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.413 |
-0.032 |
| 371 |
254485_at |
AT4G20760
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.411 |
-0.117 |
| 372 |
246505_at |
AT5G16250
|
unknown |
-0.409 |
0.024 |
| 373 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.409 |
-0.159 |
| 374 |
253978_at |
AT4G26660
|
unknown |
-0.408 |
-0.075 |
| 375 |
257750_at |
AT3G18800
|
unknown |
-0.408 |
-0.075 |
| 376 |
251155_at |
AT3G63160
|
OEP6, outer envelope protein 6 |
-0.407 |
-0.297 |
| 377 |
252039_at |
AT3G52155
|
[Phosphoglycerate mutase family protein] |
-0.404 |
-0.097 |
| 378 |
248150_at |
AT5G54670
|
ATK3, kinesin 3, KATC, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA C |
-0.404 |
-0.192 |
| 379 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
-0.404 |
-0.281 |
| 380 |
249375_at |
AT5G40730
|
AGP24, arabinogalactan protein 24, ATAGP24, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEIN 24 |
-0.404 |
0.079 |
| 381 |
251594_at |
AT3G57630
|
[exostosin family protein] |
-0.404 |
0.119 |
| 382 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.403 |
-0.279 |
| 383 |
257875_at |
AT3G17120
|
unknown |
-0.403 |
-0.058 |
| 384 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
-0.403 |
-0.156 |
| 385 |
251058_at |
AT5G01790
|
unknown |
-0.403 |
-0.393 |
| 386 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.402 |
-0.144 |
| 387 |
246387_at |
AT1G77400
|
unknown |
-0.402 |
-0.117 |
| 388 |
260739_at |
AT1G15000
|
scpl50, serine carboxypeptidase-like 50 |
-0.401 |
-0.074 |
| 389 |
263782_at |
AT2G46380
|
unknown |
-0.401 |
-0.202 |
| 390 |
267382_at |
AT2G44300
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.401 |
0.102 |
| 391 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.400 |
-0.140 |
| 392 |
247486_at |
AT5G62140
|
unknown |
-0.400 |
-0.395 |
| 393 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
-0.400 |
-0.075 |