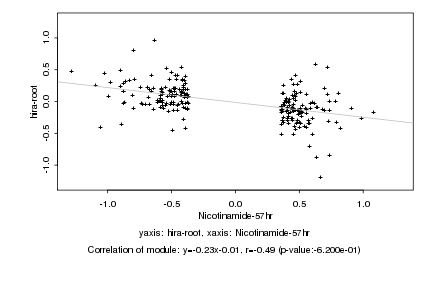
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Nicotinamide-57hr |
Log2 signal ratio
hira-root |
| 1 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
1.080 |
-0.167 |
| 2 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.979 |
-0.266 |
| 3 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.908 |
-0.105 |
| 4 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.816 |
-0.422 |
| 5 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.805 |
0.131 |
| 6 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
0.790 |
-0.326 |
| 7 |
260664_at |
AT1G19510
|
ATRL5, RAD-like 5, RL5, RAD-like 5, RSM4, RADIALIS-LIKE SANT/MYB 4 |
0.778 |
0.008 |
| 8 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.736 |
-0.833 |
| 9 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.732 |
0.009 |
| 10 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.730 |
-0.140 |
| 11 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.727 |
-0.306 |
| 12 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.718 |
0.550 |
| 13 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.714 |
0.125 |
| 14 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
0.692 |
0.215 |
| 15 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
0.690 |
-0.134 |
| 16 |
259953_at |
AT1G74810
|
BOR5, REQUIRES HIGH BORON 5 |
0.675 |
-0.123 |
| 17 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.662 |
-1.188 |
| 18 |
249454_at |
AT5G39520
|
unknown |
0.635 |
-0.085 |
| 19 |
264543_at |
AT1G55790
|
[FUNCTIONS IN: metal ion binding] |
0.631 |
-0.875 |
| 20 |
248700_at |
AT5G48400
|
ATGLR1.2, GLR1.2, GLUTAMATE RECEPTOR 1.2 |
0.628 |
-0.084 |
| 21 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
0.626 |
0.583 |
| 22 |
253182_at |
AT4G35190
|
LOG5, LONELY GUY 5 |
0.612 |
-0.025 |
| 23 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.601 |
-0.511 |
| 24 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.600 |
-0.253 |
| 25 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
0.597 |
-0.009 |
| 26 |
254276_at |
AT4G22820
|
[A20/AN1-like zinc finger family protein] |
0.586 |
-0.109 |
| 27 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
0.581 |
-0.031 |
| 28 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.578 |
-0.692 |
| 29 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.578 |
-0.332 |
| 30 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.566 |
-0.337 |
| 31 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.565 |
-0.298 |
| 32 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.556 |
0.122 |
| 33 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
0.556 |
-0.110 |
| 34 |
258752_at |
AT3G09520
|
ATEXO70H4, exocyst subunit exo70 family protein H4, EXO70H4, exocyst subunit exo70 family protein H4 |
0.554 |
-0.187 |
| 35 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.549 |
-0.399 |
| 36 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.542 |
-0.108 |
| 37 |
247532_at |
AT5G61560
|
[U-box domain-containing protein kinase family protein] |
0.536 |
-0.386 |
| 38 |
264375_at |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
0.524 |
-0.058 |
| 39 |
248701_at |
AT5G48410
|
ATGLR1.3, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 1.3, GLR1.3, glutamate receptor 1.3 |
0.519 |
-0.170 |
| 40 |
259511_at |
AT1G12520
|
ATCCS, copper chaperone for SOD1, CCS, copper chaperone for SOD1 |
0.515 |
-0.142 |
| 41 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
0.513 |
-0.226 |
| 42 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.511 |
-0.162 |
| 43 |
247212_at |
AT5G65040
|
unknown |
0.508 |
-0.161 |
| 44 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
0.506 |
-0.104 |
| 45 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.505 |
0.327 |
| 46 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.505 |
0.155 |
| 47 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
0.505 |
-0.341 |
| 48 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
0.502 |
-0.345 |
| 49 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.498 |
-0.184 |
| 50 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.497 |
-0.315 |
| 51 |
264809_at |
AT1G08830
|
CSD1, copper/zinc superoxide dismutase 1 |
0.496 |
-0.145 |
| 52 |
256818_at |
AT3G21420
|
LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1 |
0.496 |
-0.398 |
| 53 |
248896_at |
AT5G46350
|
ATWRKY8, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 8, WRKY8, WRKY DNA-binding protein 8 |
0.488 |
-0.194 |
| 54 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
0.488 |
-0.130 |
| 55 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
0.486 |
-0.199 |
| 56 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.481 |
-0.296 |
| 57 |
254014_at |
AT4G26120
|
[Ankyrin repeat family protein / BTB/POZ domain-containing protein] |
0.481 |
-0.141 |
| 58 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.481 |
0.279 |
| 59 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.478 |
-0.317 |
| 60 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
0.472 |
-0.331 |
| 61 |
262731_at |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
0.471 |
0.132 |
| 62 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
0.469 |
0.080 |
| 63 |
258323_at |
AT3G22750
|
[Protein kinase superfamily protein] |
0.468 |
0.020 |
| 64 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.467 |
0.418 |
| 65 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.466 |
-0.134 |
| 66 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
0.465 |
-0.433 |
| 67 |
259976_at |
AT1G76560
|
CP12-3, CP12 domain-containing protein 3 |
0.456 |
0.104 |
| 68 |
247282_at |
AT5G64240
|
AtMC3, metacaspase 3, AtMCP1a, metacaspase 1a, MC3, metacaspase 3, MCP1a, metacaspase 1a |
0.456 |
0.173 |
| 69 |
254710_at |
AT4G18050
|
ABCB9, ATP-binding cassette B9, PGP9, P-glycoprotein 9 |
0.456 |
-0.389 |
| 70 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.453 |
-0.148 |
| 71 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.452 |
-0.102 |
| 72 |
248011_at |
AT5G56300
|
GAMT2, gibberellic acid methyltransferase 2 |
0.449 |
-0.046 |
| 73 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.448 |
-0.506 |
| 74 |
258170_at |
AT3G21600
|
[Senescence/dehydration-associated protein-related] |
0.447 |
0.281 |
| 75 |
256574_at |
AT3G14780
|
unknown |
0.447 |
0.057 |
| 76 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
0.446 |
-0.120 |
| 77 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.446 |
0.096 |
| 78 |
265203_at |
AT2G36630
|
[Sulfite exporter TauE/SafE family protein] |
0.443 |
-0.083 |
| 79 |
251727_at |
AT3G56290
|
unknown |
0.442 |
-0.297 |
| 80 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.441 |
-0.255 |
| 81 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.438 |
-0.108 |
| 82 |
260337_at |
AT1G69310
|
ATWRKY57, WRKY57, WRKY DNA-binding protein 57 |
0.437 |
-0.265 |
| 83 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.436 |
0.350 |
| 84 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
0.426 |
-0.157 |
| 85 |
259629_at |
AT1G56510
|
ADR2, ACTIVATED DISEASE RESISTANCE 2, WRR4, WHITE RUST RESISTANCE 4 |
0.423 |
0.032 |
| 86 |
266165_at |
AT2G28190
|
CSD2, copper/zinc superoxide dismutase 2, CZSOD2, COPPER/ZINC SUPEROXIDE DISMUTASE 2 |
0.417 |
-0.048 |
| 87 |
248239_at |
AT5G53920
|
[ribosomal protein L11 methyltransferase-related] |
0.415 |
-0.003 |
| 88 |
259320_at |
AT3G01080
|
ATWRKY58, WRKY DNA-BINDING PROTEIN 58, WRKY58, WRKY DNA-binding protein 58 |
0.414 |
-0.341 |
| 89 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.413 |
-0.229 |
| 90 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.410 |
-0.176 |
| 91 |
260327_at |
AT1G63840
|
[RING/U-box superfamily protein] |
0.410 |
0.010 |
| 92 |
261866_at |
AT1G50420
|
SCL-3, SCARECROW-LIKE 3, SCL3, scarecrow-like 3 |
0.406 |
-0.093 |
| 93 |
254410_at |
AT4G21410
|
CRK29, cysteine-rich RLK (RECEPTOR-like protein kinase) 29 |
0.402 |
-0.263 |
| 94 |
256296_at |
AT1G69480
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.400 |
-0.139 |
| 95 |
245420_at |
AT4G17410
|
[DWNN domain, a CCHC-type zinc finger] |
0.400 |
-0.287 |
| 96 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
0.396 |
0.054 |
| 97 |
248080_at |
AT5G55380
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
0.394 |
-0.014 |
| 98 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.392 |
-0.153 |
| 99 |
261934_at |
AT1G22400
|
ATUGT85A1, ARABIDOPSIS THALIANA UDP-GLUCOSYL TRANSFERASE 85A1, UGT85A1 |
0.390 |
0.017 |
| 100 |
264692_at |
AT1G70000
|
[myb-like transcription factor family protein] |
0.379 |
-0.005 |
| 101 |
247956_at |
AT5G56970
|
ATCKX3, CKX3, cytokinin oxidase 3 |
0.377 |
-0.057 |
| 102 |
249622_at |
AT5G37550
|
unknown |
0.375 |
-0.046 |
| 103 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.375 |
-0.322 |
| 104 |
261973_at |
AT1G64610
|
[Transducin/WD40 repeat-like superfamily protein] |
0.374 |
-0.247 |
| 105 |
258468_at |
AT3G06070
|
unknown |
0.371 |
0.253 |
| 106 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
0.370 |
0.133 |
| 107 |
254797_at |
AT4G13030
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.367 |
-0.120 |
| 108 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
0.365 |
0.138 |
| 109 |
266269_at |
AT2G29480
|
ATGSTU2, glutathione S-transferase tau 2, GST20, GLUTATHIONE S-TRANSFERASE 20, GSTU2, glutathione S-transferase tau 2 |
0.362 |
-0.285 |
| 110 |
267226_at |
AT2G44010
|
unknown |
0.362 |
-0.155 |
| 111 |
256321_at |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
0.361 |
-0.285 |
| 112 |
250723_at |
AT5G06300
|
LOG7, LONELY GUY 7 |
0.360 |
-0.066 |
| 113 |
265659_at |
AT2G25440
|
AtRLP20, receptor like protein 20, RLP20, receptor like protein 20 |
0.360 |
-0.068 |
| 114 |
253453_at |
AT4G31860
|
[Protein phosphatase 2C family protein] |
0.359 |
-0.131 |
| 115 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.358 |
-0.124 |
| 116 |
259970_at |
AT1G76570
|
AtLHCB7, LHCB7, light-harvesting complex B7 |
0.355 |
-0.159 |
| 117 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
0.354 |
-0.517 |
| 118 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.353 |
-0.352 |
| 119 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-1.289 |
0.487 |
| 120 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-1.102 |
0.260 |
| 121 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
-1.058 |
-0.401 |
| 122 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-1.031 |
0.456 |
| 123 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
-0.999 |
0.093 |
| 124 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.985 |
0.311 |
| 125 |
250778_at |
AT5G05500
|
MOP10 |
-0.904 |
0.243 |
| 126 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.903 |
0.498 |
| 127 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-0.897 |
-0.360 |
| 128 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-0.884 |
0.299 |
| 129 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.882 |
0.161 |
| 130 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.881 |
0.164 |
| 131 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
-0.878 |
-0.022 |
| 132 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.876 |
-0.013 |
| 133 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.868 |
0.321 |
| 134 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.830 |
0.335 |
| 135 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.808 |
0.103 |
| 136 |
265050_at |
AT1G52070
|
[Mannose-binding lectin superfamily protein] |
-0.805 |
0.816 |
| 137 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
-0.800 |
-0.102 |
| 138 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.795 |
0.358 |
| 139 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-0.745 |
0.221 |
| 140 |
249545_at |
AT5G38030
|
[MATE efflux family protein] |
-0.743 |
-0.019 |
| 141 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.735 |
-0.041 |
| 142 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
-0.709 |
-0.038 |
| 143 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.695 |
0.221 |
| 144 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.683 |
0.065 |
| 145 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
-0.680 |
-0.039 |
| 146 |
267459_at |
AT2G33850
|
unknown |
-0.674 |
0.194 |
| 147 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
-0.663 |
0.416 |
| 148 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.660 |
0.158 |
| 149 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.649 |
0.219 |
| 150 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.644 |
-0.118 |
| 151 |
261691_at |
AT1G50060
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.635 |
0.960 |
| 152 |
262373_at |
AT1G73120
|
unknown |
-0.613 |
-0.009 |
| 153 |
265248_at |
AT2G43010
|
AtPIF4, PIF4, phytochrome interacting factor 4, SRL2 |
-0.611 |
0.031 |
| 154 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.595 |
0.016 |
| 155 |
266941_at |
AT2G18980
|
[Peroxidase superfamily protein] |
-0.593 |
0.097 |
| 156 |
264987_at |
AT1G27030
|
unknown |
-0.591 |
0.006 |
| 157 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
-0.589 |
-0.081 |
| 158 |
253285_at |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
-0.588 |
0.051 |
| 159 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
-0.586 |
0.190 |
| 160 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
-0.586 |
0.166 |
| 161 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
-0.581 |
0.208 |
| 162 |
253244_at |
AT4G34580
|
AtSFH1, COW1, CAN OF WORMS1, SRH1, SHORT ROOT HAIR 1 |
-0.578 |
0.149 |
| 163 |
253317_at |
AT4G33960
|
unknown |
-0.575 |
0.115 |
| 164 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.574 |
-0.061 |
| 165 |
253947_at |
AT4G26760
|
MAP65-2, microtubule-associated protein 65-2 |
-0.572 |
0.116 |
| 166 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.572 |
-0.000 |
| 167 |
262598_at |
AT1G15260
|
unknown |
-0.571 |
0.017 |
| 168 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
-0.569 |
-0.094 |
| 169 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.553 |
0.224 |
| 170 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.552 |
-0.059 |
| 171 |
252149_at |
AT3G51290
|
APSR1, Altered Phosphate Starvation Response 1 |
-0.547 |
-0.021 |
| 172 |
259996_at |
AT1G67910
|
unknown |
-0.541 |
0.520 |
| 173 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.535 |
0.085 |
| 174 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.529 |
-0.146 |
| 175 |
255876_at |
AT2G40480
|
unknown |
-0.523 |
0.350 |
| 176 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.521 |
0.177 |
| 177 |
255460_at |
AT4G02800
|
unknown |
-0.519 |
0.116 |
| 178 |
248252_at |
AT5G53250
|
AGP22, arabinogalactan protein 22, ATAGP22, ARABINOGALACTAN PROTEIN 22 |
-0.514 |
0.159 |
| 179 |
253729_at |
AT4G29360
|
[O-Glycosyl hydrolases family 17 protein] |
-0.510 |
-0.038 |
| 180 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
-0.508 |
0.469 |
| 181 |
265083_at |
AT1G03820
|
unknown |
-0.507 |
0.167 |
| 182 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.504 |
-0.003 |
| 183 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
-0.503 |
0.083 |
| 184 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.494 |
-0.451 |
| 185 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.494 |
0.114 |
| 186 |
257115_at |
AT3G20150
|
[Kinesin motor family protein] |
-0.490 |
0.205 |
| 187 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-0.489 |
-0.129 |
| 188 |
266089_at |
AT2G38010
|
[Neutral/alkaline non-lysosomal ceramidase] |
-0.488 |
-0.011 |
| 189 |
247239_at |
AT5G64640
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.486 |
-0.046 |
| 190 |
263902_at |
AT2G36230
|
APG10, ALBINO AND PALE GREEN 10, HISN3 |
-0.481 |
0.086 |
| 191 |
250059_at |
AT5G17820
|
[Peroxidase superfamily protein] |
-0.480 |
0.209 |
| 192 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
-0.479 |
0.170 |
| 193 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.474 |
0.218 |
| 194 |
246565_at |
AT5G15530
|
BCCP2, biotin carboxyl carrier protein 2, CAC1-B |
-0.473 |
0.413 |
| 195 |
249167_at |
AT5G42860
|
unknown |
-0.468 |
0.112 |
| 196 |
267264_at |
AT2G22970
|
SCPL11, serine carboxypeptidase-like 11 |
-0.466 |
0.356 |
| 197 |
251142_at |
AT5G01015
|
unknown |
-0.461 |
-0.132 |
| 198 |
252995_at |
AT4G38370
|
[Phosphoglycerate mutase family protein] |
-0.456 |
-0.056 |
| 199 |
266336_at |
AT2G32270
|
ZIP3, zinc transporter 3 precursor |
-0.454 |
0.419 |
| 200 |
247603_at |
AT5G60930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.448 |
-0.037 |
| 201 |
261780_at |
AT1G76310
|
CYCB2;4, CYCLIN B2;4 |
-0.444 |
0.201 |
| 202 |
249002_at |
AT5G44520
|
[NagB/RpiA/CoA transferase-like superfamily protein] |
-0.432 |
0.095 |
| 203 |
264802_at |
AT1G08560
|
ATSYP111, KN, KNOLLE, SYP111, syntaxin of plants 111 |
-0.430 |
0.152 |
| 204 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
-0.428 |
0.542 |
| 205 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
-0.426 |
0.103 |
| 206 |
264319_at |
AT1G04110
|
AtSDD1, SDD1, STOMATAL DENSITY AND DISTRIBUTION 1 |
-0.424 |
0.012 |
| 207 |
250560_at |
AT5G08020
|
ATRPA70B, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT B, RPA70B, RPA70-kDa subunit B |
-0.423 |
0.172 |
| 208 |
245930_at |
AT5G09240
|
[ssDNA-binding transcriptional regulator] |
-0.420 |
0.143 |
| 209 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
-0.420 |
-0.025 |
| 210 |
254573_at |
AT4G19420
|
[Pectinacetylesterase family protein] |
-0.420 |
0.336 |
| 211 |
259358_at |
AT1G13250
|
GATL3, galacturonosyltransferase-like 3 |
-0.417 |
0.359 |
| 212 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.414 |
-0.090 |
| 213 |
253613_at |
AT4G30320
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.411 |
0.235 |
| 214 |
258067_at |
AT3G25980
|
AtMAD2, MAD2, MITOTIC ARREST-DEFICIENT 2 |
-0.411 |
0.204 |
| 215 |
258618_at |
AT3G02885
|
GASA5, GAST1 protein homolog 5 |
-0.410 |
-0.267 |
| 216 |
258633_at |
AT3G07990
|
SCPL27, serine carboxypeptidase-like 27 |
-0.408 |
0.254 |
| 217 |
246439_at |
AT5G17600
|
[RING/U-box superfamily protein] |
-0.406 |
0.132 |
| 218 |
256052_at |
AT1G06960
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.403 |
0.077 |
| 219 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.400 |
0.341 |
| 220 |
258151_at |
AT3G18080
|
BGLU44, B-S glucosidase 44 |
-0.398 |
0.291 |
| 221 |
260353_at |
AT1G69230
|
SP1L2, SPIRAL1-like2 |
-0.397 |
0.098 |
| 222 |
257516_at |
AT1G69040
|
ACR4, ACT domain repeat 4 |
-0.396 |
-0.024 |
| 223 |
266489_at |
AT2G35190
|
ATNPSN11, NPSN11, novel plant snare 11, NSPN11 |
-0.396 |
0.193 |
| 224 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-0.394 |
-0.100 |
| 225 |
249046_at |
AT5G44400
|
[FAD-binding Berberine family protein] |
-0.393 |
-0.424 |
| 226 |
267191_at |
AT2G44110
|
ATMLO15, MILDEW RESISTANCE LOCUS O 15, MLO15, MILDEW RESISTANCE LOCUS O 15 |
-0.393 |
0.400 |
| 227 |
258006_at |
AT3G19400
|
[Cysteine proteinases superfamily protein] |
-0.388 |
0.123 |
| 228 |
251261_at |
AT3G62110
|
[Pectin lyase-like superfamily protein] |
-0.382 |
0.073 |
| 229 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.382 |
-0.013 |
| 230 |
245828_at |
AT1G57820
|
ORTH2, ORTHRUS 2, VIM1, VARIANT IN METHYLATION 1 |
-0.381 |
-0.119 |
| 231 |
263120_at |
AT1G78490
|
CYP708A3, cytochrome P450, family 708, subfamily A, polypeptide 3 |
-0.381 |
-0.106 |
| 232 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.378 |
0.192 |
| 233 |
253403_at |
AT4G32830
|
AtAUR1, ataurora1, AUR1, ataurora1 |
-0.376 |
0.112 |
| 234 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
-0.375 |
-0.016 |
| 235 |
252039_at |
AT3G52155
|
[Phosphoglycerate mutase family protein] |
-0.375 |
0.086 |