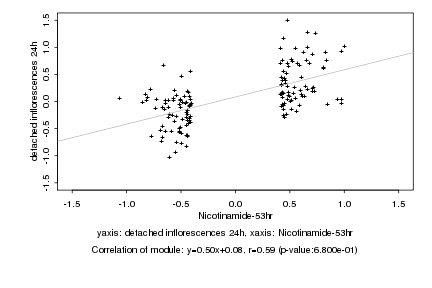
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Nicotinamide-53hr |
Log2 signal ratio
detached inflorescences 24h |
| 1 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
0.999 |
1.020 |
| 2 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.974 |
0.938 |
| 3 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
0.972 |
-0.030 |
| 4 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.968 |
0.049 |
| 5 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.929 |
0.042 |
| 6 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.845 |
-0.040 |
| 7 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.835 |
0.775 |
| 8 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.825 |
0.922 |
| 9 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.807 |
0.631 |
| 10 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.801 |
0.616 |
| 11 |
249454_at |
AT5G39520
|
unknown |
0.733 |
1.262 |
| 12 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.725 |
0.189 |
| 13 |
248563_at |
AT5G49690
|
[UDP-Glycosyltransferase superfamily protein] |
0.711 |
0.271 |
| 14 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
0.707 |
0.243 |
| 15 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.705 |
0.200 |
| 16 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.701 |
0.883 |
| 17 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.672 |
0.715 |
| 18 |
263475_at |
AT2G31945
|
unknown |
0.662 |
1.276 |
| 19 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.659 |
0.234 |
| 20 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.653 |
0.998 |
| 21 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.651 |
0.764 |
| 22 |
267036_at |
AT2G38465
|
unknown |
0.635 |
0.294 |
| 23 |
254014_at |
AT4G26120
|
[Ankyrin repeat family protein / BTB/POZ domain-containing protein] |
0.628 |
0.098 |
| 24 |
260744_at |
AT1G15010
|
unknown |
0.624 |
0.918 |
| 25 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.615 |
0.111 |
| 26 |
258813_at |
AT3G04060
|
anac046, NAC domain containing protein 46, NAC046, NAC domain containing protein 46 |
0.606 |
0.461 |
| 27 |
247205_at |
AT5G64890
|
PROPEP2, elicitor peptide 2 precursor |
0.603 |
0.147 |
| 28 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
0.595 |
0.204 |
| 29 |
251647_at |
AT3G57770
|
[Protein kinase superfamily protein] |
0.586 |
0.677 |
| 30 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
0.583 |
-0.059 |
| 31 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
0.567 |
0.707 |
| 32 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.553 |
-0.179 |
| 33 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.548 |
0.996 |
| 34 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.547 |
0.067 |
| 35 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.534 |
0.273 |
| 36 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
0.533 |
0.160 |
| 37 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.522 |
0.756 |
| 38 |
259511_at |
AT1G12520
|
ATCCS, copper chaperone for SOD1, CCS, copper chaperone for SOD1 |
0.509 |
0.025 |
| 39 |
264809_at |
AT1G08830
|
CSD1, copper/zinc superoxide dismutase 1 |
0.509 |
-0.135 |
| 40 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.507 |
0.783 |
| 41 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
0.500 |
0.017 |
| 42 |
253322_at |
AT4G33980
|
unknown |
0.490 |
0.098 |
| 43 |
264082_at |
AT2G28570
|
unknown |
0.487 |
0.168 |
| 44 |
257950_at |
AT3G21780
|
UGT71B6, UDP-glucosyl transferase 71B6 |
0.482 |
0.124 |
| 45 |
253827_at |
AT4G28085
|
unknown |
0.480 |
0.657 |
| 46 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
0.476 |
0.711 |
| 47 |
252977_at |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.475 |
0.046 |
| 48 |
261934_at |
AT1G22400
|
ATUGT85A1, ARABIDOPSIS THALIANA UDP-GLUCOSYL TRANSFERASE 85A1, UGT85A1 |
0.471 |
0.294 |
| 49 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.471 |
1.513 |
| 50 |
251621_at |
AT3G57700
|
[Protein kinase superfamily protein] |
0.463 |
0.527 |
| 51 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
0.461 |
-0.234 |
| 52 |
256839_at |
AT3G22930
|
AtCML11, CML11, calmodulin-like 11 |
0.454 |
0.389 |
| 53 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
0.451 |
0.347 |
| 54 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.450 |
-0.283 |
| 55 |
245705_at |
AT5G04390
|
[C2H2-type zinc finger family protein] |
0.448 |
0.427 |
| 56 |
248934_at |
AT5G46080
|
[Protein kinase superfamily protein] |
0.444 |
-0.026 |
| 57 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.441 |
-0.066 |
| 58 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
0.438 |
0.557 |
| 59 |
259945_at |
AT1G71460
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.437 |
-0.245 |
| 60 |
264288_at |
AT1G62045
|
unknown |
0.437 |
-0.135 |
| 61 |
249705_at |
AT5G35580
|
[Protein kinase superfamily protein] |
0.435 |
0.162 |
| 62 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.433 |
1.170 |
| 63 |
258110_at |
AT3G14610
|
CYP72A7, cytochrome P450, family 72, subfamily A, polypeptide 7 |
0.432 |
0.167 |
| 64 |
260030_at |
AT1G68880
|
AtbZIP, basic leucine-zipper 8, bZIP, basic leucine-zipper 8 |
0.431 |
0.086 |
| 65 |
247177_at |
AT5G65300
|
unknown |
0.431 |
-0.039 |
| 66 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
0.430 |
0.763 |
| 67 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
0.428 |
0.396 |
| 68 |
247282_at |
AT5G64240
|
AtMC3, metacaspase 3, AtMCP1a, metacaspase 1a, MC3, metacaspase 3, MCP1a, metacaspase 1a |
0.423 |
0.458 |
| 69 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.421 |
-0.091 |
| 70 |
265926_at |
AT2G18600
|
[Ubiquitin-conjugating enzyme family protein] |
0.420 |
0.144 |
| 71 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.418 |
0.328 |
| 72 |
260337_at |
AT1G69310
|
ATWRKY57, WRKY57, WRKY DNA-binding protein 57 |
0.416 |
0.152 |
| 73 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.415 |
0.305 |
| 74 |
258323_at |
AT3G22750
|
[Protein kinase superfamily protein] |
0.414 |
0.707 |
| 75 |
244974_at |
ATCG00700
|
PSBN, photosystem II reaction center protein N |
0.411 |
0.132 |
| 76 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.411 |
0.990 |
| 77 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-1.069 |
0.070 |
| 78 |
265063_at |
AT1G61500
|
[S-locus lectin protein kinase family protein] |
-0.863 |
-0.001 |
| 79 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.832 |
0.134 |
| 80 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.822 |
0.030 |
| 81 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.815 |
0.091 |
| 82 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.784 |
0.238 |
| 83 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-0.776 |
-0.629 |
| 84 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.735 |
-0.124 |
| 85 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.731 |
0.055 |
| 86 |
258341_at |
AT3G22790
|
NET1A, Networked 1A |
-0.693 |
-0.532 |
| 87 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-0.684 |
-0.727 |
| 88 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.675 |
-0.446 |
| 89 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.674 |
-0.109 |
| 90 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
-0.673 |
-0.653 |
| 91 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.662 |
0.665 |
| 92 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
-0.659 |
-0.138 |
| 93 |
262598_at |
AT1G15260
|
unknown |
-0.651 |
-0.023 |
| 94 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.651 |
-0.537 |
| 95 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
-0.646 |
0.031 |
| 96 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
-0.623 |
-0.109 |
| 97 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.621 |
0.023 |
| 98 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.619 |
-0.294 |
| 99 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.614 |
-0.223 |
| 100 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.610 |
-1.029 |
| 101 |
246415_at |
AT5G17160
|
unknown |
-0.591 |
-0.538 |
| 102 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
-0.579 |
-0.242 |
| 103 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.577 |
0.023 |
| 104 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.574 |
0.065 |
| 105 |
263902_at |
AT2G36230
|
APG10, ALBINO AND PALE GREEN 10, HISN3 |
-0.569 |
-0.365 |
| 106 |
259091_at |
AT3G04890
|
[Uncharacterized conserved protein (DUF2358)] |
-0.566 |
0.221 |
| 107 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.557 |
-0.924 |
| 108 |
248057_at |
AT5G55520
|
unknown |
-0.545 |
-0.741 |
| 109 |
247603_at |
AT5G60930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.543 |
-0.264 |
| 110 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.542 |
0.112 |
| 111 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
-0.523 |
-0.554 |
| 112 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.521 |
-0.054 |
| 113 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
-0.517 |
-0.483 |
| 114 |
256864_at |
AT3G23890
|
ATTOPII, TOPII, topoisomerase II |
-0.516 |
-0.542 |
| 115 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.514 |
-0.098 |
| 116 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-0.513 |
0.045 |
| 117 |
266009_at |
AT2G37420
|
[ATP binding microtubule motor family protein] |
-0.506 |
-0.470 |
| 118 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
-0.505 |
0.021 |
| 119 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.504 |
0.474 |
| 120 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
-0.503 |
-0.586 |
| 121 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.502 |
-0.770 |
| 122 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
-0.493 |
-0.002 |
| 123 |
253788_at |
AT4G28680
|
AtTYDC, TYDC, L-tyrosine decarboxylase, TYRDC, L-tyrosine decarboxylase, TYRDC1, L-TYROSINE DECARBOXYLASE 1 |
-0.489 |
-0.320 |
| 124 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.472 |
0.107 |
| 125 |
249002_at |
AT5G44520
|
[NagB/RpiA/CoA transferase-like superfamily protein] |
-0.467 |
-0.036 |
| 126 |
255410_at |
AT4G03100
|
[Rho GTPase activating protein with PAK-box/P21-Rho-binding domain] |
-0.457 |
-0.620 |
| 127 |
253729_at |
AT4G29360
|
[O-Glycosyl hydrolases family 17 protein] |
-0.454 |
-0.196 |
| 128 |
255876_at |
AT2G40480
|
unknown |
-0.454 |
-0.427 |
| 129 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.453 |
-0.279 |
| 130 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.452 |
-0.012 |
| 131 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.450 |
-0.826 |
| 132 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
-0.449 |
-0.232 |
| 133 |
262651_at |
AT1G14100
|
FUT8, fucosyltransferase 8 |
-0.448 |
-0.346 |
| 134 |
247028_at |
AT5G67100
|
ICU2, INCURVATA2 |
-0.446 |
-0.630 |
| 135 |
255851_at |
AT1G67040
|
TRM22, TON1 Recruiting Motif 22 |
-0.444 |
-0.157 |
| 136 |
258471_at |
AT3G06030
|
ANP3, NPK1-related protein kinase 3, AtANP3, MAPKKK12, NP3, NPK1-related protein kinase 3 |
-0.442 |
-0.622 |
| 137 |
257761_at |
AT3G23090
|
WDL3 |
-0.441 |
0.191 |
| 138 |
250381_at |
AT5G11610
|
[Exostosin family protein] |
-0.440 |
0.181 |
| 139 |
245930_at |
AT5G09240
|
[ssDNA-binding transcriptional regulator] |
-0.433 |
-0.397 |
| 140 |
265083_at |
AT1G03820
|
unknown |
-0.431 |
-0.302 |
| 141 |
266330_at |
AT2G01530
|
MLP329, MLP-like protein 329, ZCE2, (Zusammen-CA)-enhanced 2 |
-0.429 |
0.096 |
| 142 |
253244_at |
AT4G34580
|
AtSFH1, COW1, CAN OF WORMS1, SRH1, SHORT ROOT HAIR 1 |
-0.426 |
-0.333 |
| 143 |
245575_at |
AT4G14760
|
NET1B, Networked 1B |
-0.425 |
-0.078 |
| 144 |
253254_at |
AT4G34650
|
SQS2, squalene synthase 2 |
-0.425 |
-0.072 |
| 145 |
247452_at |
AT5G62430
|
CDF1, cycling DOF factor 1 |
-0.421 |
0.045 |
| 146 |
256585_at |
AT3G28760
|
unknown |
-0.420 |
-0.370 |
| 147 |
247097_at |
AT5G66460
|
AtMAN7, MAN7, endo-beta-mannase 7 |
-0.418 |
-0.268 |
| 148 |
252089_at |
AT3G52110
|
unknown |
-0.417 |
-0.071 |
| 149 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
-0.417 |
-0.048 |
| 150 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
-0.416 |
0.557 |
| 151 |
263687_at |
AT1G26940
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.406 |
-0.025 |