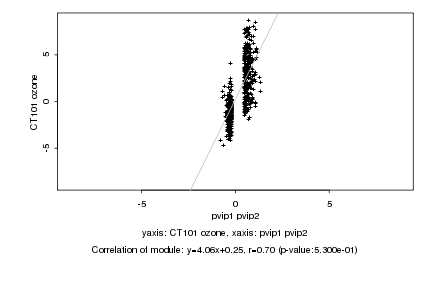
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
pvip1 pvip2 |
Log2 signal ratio
CT101 ozone |
| 1 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.302 |
1.177 |
| 2 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
1.284 |
2.114 |
| 3 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
1.261 |
2.607 |
| 4 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
1.160 |
5.287 |
| 5 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
1.101 |
4.716 |
| 6 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
1.091 |
5.591 |
| 7 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
1.077 |
5.723 |
| 8 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.062 |
-0.027 |
| 9 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
1.061 |
4.547 |
| 10 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
1.049 |
5.456 |
| 11 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
1.045 |
-0.455 |
| 12 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
1.035 |
8.498 |
| 13 |
252511_at |
AT3G46280
|
[protein kinase-related] |
1.031 |
2.164 |
| 14 |
246366_at |
AT1G51850
|
[Leucine-rich repeat protein kinase family protein] |
1.029 |
3.205 |
| 15 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
1.028 |
7.752 |
| 16 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
1.023 |
2.900 |
| 17 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
1.019 |
-0.177 |
| 18 |
259385_at |
AT1G13470
|
unknown |
1.010 |
2.884 |
| 19 |
264635_at |
AT1G65500
|
unknown |
0.985 |
2.361 |
| 20 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.968 |
2.978 |
| 21 |
258203_at |
AT3G13950
|
unknown |
0.963 |
5.327 |
| 22 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.961 |
8.076 |
| 23 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.941 |
2.267 |
| 24 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
0.929 |
1.356 |
| 25 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.920 |
7.066 |
| 26 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.912 |
6.216 |
| 27 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.909 |
0.483 |
| 28 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.906 |
2.730 |
| 29 |
253024_at |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
0.893 |
0.386 |
| 30 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.892 |
4.483 |
| 31 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
0.891 |
0.245 |
| 32 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
0.889 |
-0.051 |
| 33 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
0.881 |
1.943 |
| 34 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
0.879 |
3.252 |
| 35 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
0.876 |
4.703 |
| 36 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.875 |
4.569 |
| 37 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.859 |
3.436 |
| 38 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.854 |
0.774 |
| 39 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.848 |
4.596 |
| 40 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.847 |
6.988 |
| 41 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.844 |
6.595 |
| 42 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.843 |
2.374 |
| 43 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.838 |
0.088 |
| 44 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
0.837 |
1.424 |
| 45 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.834 |
0.222 |
| 46 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
0.831 |
2.079 |
| 47 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
0.830 |
4.350 |
| 48 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.828 |
2.232 |
| 49 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.817 |
4.227 |
| 50 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.813 |
3.607 |
| 51 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
0.802 |
3.593 |
| 52 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.801 |
4.413 |
| 53 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.801 |
5.591 |
| 54 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.800 |
3.857 |
| 55 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
0.799 |
4.642 |
| 56 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
0.798 |
5.713 |
| 57 |
267121_at |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.795 |
-0.638 |
| 58 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.794 |
1.466 |
| 59 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.789 |
5.282 |
| 60 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.780 |
4.145 |
| 61 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.780 |
2.090 |
| 62 |
261684_at |
AT1G47400
|
unknown |
0.768 |
-0.150 |
| 63 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.767 |
2.513 |
| 64 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
0.762 |
0.920 |
| 65 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.759 |
2.925 |
| 66 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.758 |
3.550 |
| 67 |
251895_at |
AT3G54420
|
ATCHITIV, CHITINASE CLASS IV, ATEP3, homolog of carrot EP3-3 chitinase, CHIV, EP3, homolog of carrot EP3-3 chitinase |
0.755 |
5.013 |
| 68 |
249032_at |
AT5G44910
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.755 |
2.562 |
| 69 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.753 |
4.789 |
| 70 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.746 |
3.169 |
| 71 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.739 |
-1.610 |
| 72 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.739 |
4.211 |
| 73 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.737 |
4.414 |
| 74 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.737 |
7.945 |
| 75 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.735 |
1.924 |
| 76 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.735 |
7.267 |
| 77 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.733 |
3.692 |
| 78 |
260005_at |
AT1G67920
|
unknown |
0.733 |
5.166 |
| 79 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.725 |
6.685 |
| 80 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.723 |
4.642 |
| 81 |
266761_at |
AT2G47130
|
AtSDR3, SDR3, short-chain dehydrogenase/reductase 2 |
0.721 |
3.296 |
| 82 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
0.713 |
1.753 |
| 83 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.709 |
0.149 |
| 84 |
262260_at |
AT1G70850
|
MLP34, MLP-like protein 34 |
0.708 |
-0.212 |
| 85 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
0.706 |
1.650 |
| 86 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.703 |
4.300 |
| 87 |
255479_at |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
0.702 |
6.209 |
| 88 |
248060_at |
AT5G55560
|
[Protein kinase superfamily protein] |
0.700 |
3.455 |
| 89 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.697 |
5.033 |
| 90 |
249454_at |
AT5G39520
|
unknown |
0.696 |
-0.794 |
| 91 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.696 |
3.783 |
| 92 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.694 |
2.521 |
| 93 |
265334_at |
AT2G18370
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.694 |
0.297 |
| 94 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.692 |
0.841 |
| 95 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
0.691 |
3.621 |
| 96 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.689 |
1.790 |
| 97 |
263182_at |
AT1G05575
|
unknown |
0.688 |
5.832 |
| 98 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.688 |
4.884 |
| 99 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
0.687 |
4.529 |
| 100 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
0.684 |
1.707 |
| 101 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
0.684 |
5.301 |
| 102 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
0.683 |
0.499 |
| 103 |
245181_at |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
0.682 |
5.387 |
| 104 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.681 |
-1.881 |
| 105 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
0.679 |
4.102 |
| 106 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.677 |
3.671 |
| 107 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.676 |
2.981 |
| 108 |
246620_at |
AT5G36220
|
CYP81D1, cytochrome P450, family 81, subfamily D, polypeptide 1, CYP91A1, CYTOCHROME P450 91A1 |
0.671 |
1.685 |
| 109 |
255596_at |
AT4G01720
|
AtWRKY47, WRKY47 |
0.670 |
1.653 |
| 110 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.670 |
0.216 |
| 111 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.669 |
0.426 |
| 112 |
264580_at |
AT1G05340
|
unknown |
0.669 |
3.696 |
| 113 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.663 |
1.814 |
| 114 |
248321_at |
AT5G52740
|
[Copper transport protein family] |
0.661 |
2.839 |
| 115 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.658 |
7.728 |
| 116 |
251531_at |
AT3G58550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.658 |
0.077 |
| 117 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.657 |
8.784 |
| 118 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.657 |
6.075 |
| 119 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.655 |
6.040 |
| 120 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.654 |
3.660 |
| 121 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.654 |
1.406 |
| 122 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
0.654 |
3.168 |
| 123 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.654 |
7.497 |
| 124 |
248298_at |
AT5G53110
|
[RING/U-box superfamily protein] |
0.652 |
3.161 |
| 125 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.652 |
7.414 |
| 126 |
264663_at |
AT1G09970
|
LRR XI-23, RLK7, receptor-like kinase 7 |
0.649 |
4.323 |
| 127 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.649 |
2.666 |
| 128 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
0.646 |
5.901 |
| 129 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.645 |
3.982 |
| 130 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
0.645 |
3.063 |
| 131 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
0.642 |
1.581 |
| 132 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
0.642 |
4.759 |
| 133 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.641 |
1.765 |
| 134 |
254252_at |
AT4G23310
|
CRK23, cysteine-rich RLK (RECEPTOR-like protein kinase) 23 |
0.639 |
2.885 |
| 135 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
0.639 |
6.198 |
| 136 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.636 |
3.012 |
| 137 |
266993_at |
AT2G39210
|
[Major facilitator superfamily protein] |
0.636 |
3.588 |
| 138 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.633 |
-0.971 |
| 139 |
266231_at |
AT2G02220
|
ATPSKR1, PHYTOSULFOKIN RECEPTOR 1, PSKR1, phytosulfokin receptor 1 |
0.627 |
3.156 |
| 140 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.626 |
5.469 |
| 141 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.625 |
5.271 |
| 142 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.624 |
5.819 |
| 143 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
0.623 |
0.652 |
| 144 |
259479_at |
AT1G19020
|
unknown |
0.621 |
7.549 |
| 145 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.615 |
7.129 |
| 146 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.611 |
2.656 |
| 147 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
0.610 |
4.605 |
| 148 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.609 |
5.306 |
| 149 |
266782_at |
AT2G29120
|
ATGLR2.7, glutamate receptor 2.7, GLR2.7, GLUTAMATE RECEPTOR 2.7, GLR2.7, glutamate receptor 2.7 |
0.608 |
1.994 |
| 150 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
0.608 |
7.908 |
| 151 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.608 |
2.676 |
| 152 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
0.606 |
6.046 |
| 153 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
0.600 |
6.242 |
| 154 |
256442_at |
AT3G10930
|
unknown |
0.600 |
3.659 |
| 155 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
0.598 |
2.039 |
| 156 |
258209_at |
AT3G14060
|
unknown |
0.598 |
0.629 |
| 157 |
264851_at |
AT2G17290
|
ATCDPK3, ARABIDOPSIS THALIANA CALMODULIN-DOMAIN PROTEIN KINASE 3, ATCPK6, ARABIDOPSIS THALIANA CALCIUM-DEPENDENT PROTEIN KINASE 6, CPK6, calcium dependent protein kinase 6 |
0.597 |
3.172 |
| 158 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.597 |
5.336 |
| 159 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
0.596 |
-0.773 |
| 160 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.596 |
4.512 |
| 161 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
0.595 |
4.378 |
| 162 |
245889_at |
AT5G09480
|
[hydroxyproline-rich glycoprotein family protein] |
0.595 |
0.979 |
| 163 |
264590_at |
AT2G17710
|
unknown |
0.592 |
0.933 |
| 164 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.589 |
7.037 |
| 165 |
267178_at |
AT2G37750
|
unknown |
0.587 |
1.830 |
| 166 |
256378_at |
AT1G66830
|
[Leucine-rich repeat protein kinase family protein] |
0.586 |
0.244 |
| 167 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.585 |
7.672 |
| 168 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.584 |
7.915 |
| 169 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.580 |
1.858 |
| 170 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
0.580 |
3.881 |
| 171 |
251304_at |
AT3G61990
|
OMTF3, O-MTase family 3 protein |
0.577 |
-0.539 |
| 172 |
266464_at |
AT2G47800
|
ABCC4, ATP-binding cassette C4, ATMRP4, multidrug resistance-associated protein 4, EST3, MRP4, multidrug resistance-associated protein 4 |
0.576 |
1.198 |
| 173 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.575 |
3.852 |
| 174 |
254524_at |
AT4G20000
|
[VQ motif-containing protein] |
0.574 |
5.263 |
| 175 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
0.570 |
0.032 |
| 176 |
261413_at |
AT1G07630
|
PLL5, pol-like 5 |
0.567 |
2.504 |
| 177 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.567 |
-0.866 |
| 178 |
266247_at |
AT2G27660
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.567 |
3.375 |
| 179 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
0.566 |
-0.151 |
| 180 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.566 |
-0.912 |
| 181 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
0.563 |
3.218 |
| 182 |
247333_at |
AT5G63600
|
ATFLS5, FLS5, flavonol synthase 5 |
0.562 |
1.543 |
| 183 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.561 |
5.845 |
| 184 |
264757_at |
AT1G61360
|
[S-locus lectin protein kinase family protein] |
0.561 |
5.396 |
| 185 |
254253_at |
AT4G23320
|
CRK24, cysteine-rich RLK (RECEPTOR-like protein kinase) 24 |
0.560 |
1.048 |
| 186 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.559 |
1.914 |
| 187 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.556 |
-0.413 |
| 188 |
249061_at |
AT5G44550
|
[Uncharacterised protein family (UPF0497)] |
0.556 |
0.229 |
| 189 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.553 |
4.724 |
| 190 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
0.552 |
0.112 |
| 191 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
0.549 |
5.521 |
| 192 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.546 |
6.916 |
| 193 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.546 |
-0.931 |
| 194 |
265720_at |
AT2G40110
|
[Yippee family putative zinc-binding protein] |
0.545 |
1.354 |
| 195 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
0.544 |
4.616 |
| 196 |
261198_at |
AT1G12940
|
ATNRT2.5, nitrate transporter2.5, NRT2.5, nitrate transporter2.5 |
0.540 |
2.751 |
| 197 |
267307_at |
AT2G30210
|
LAC3, laccase 3 |
0.536 |
-0.568 |
| 198 |
260234_at |
AT1G74460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.536 |
0.018 |
| 199 |
248306_at |
AT5G52830
|
ATWRKY27, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 27, WRKY27, WRKY DNA-binding protein 27 |
0.534 |
-0.897 |
| 200 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.533 |
-0.623 |
| 201 |
267300_at |
AT2G30140
|
UGT87A2, UDP-glucosyl transferase 87A2 |
0.533 |
2.039 |
| 202 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.532 |
1.883 |
| 203 |
260668_at |
AT1G19530
|
unknown |
0.530 |
2.917 |
| 204 |
249377_at |
AT5G40690
|
unknown |
0.529 |
1.571 |
| 205 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
0.529 |
1.345 |
| 206 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.529 |
-0.777 |
| 207 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
0.527 |
4.157 |
| 208 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.526 |
-1.048 |
| 209 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.526 |
-0.900 |
| 210 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.524 |
4.792 |
| 211 |
266884_at |
AT2G44790
|
UCC2, uclacyanin 2 |
0.524 |
4.621 |
| 212 |
259327_at |
AT3G16460
|
JAL34, jacalin-related lectin 34 |
0.520 |
-1.170 |
| 213 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
0.520 |
-0.791 |
| 214 |
248237_at |
AT5G53890
|
AtPSKR2, PSKR2, phytosylfokine-alpha receptor 2 |
0.518 |
-0.302 |
| 215 |
246098_at |
AT5G20400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.518 |
3.210 |
| 216 |
255319_at |
AT4G04220
|
AtRLP46, receptor like protein 46, RLP46, receptor like protein 46 |
0.518 |
1.422 |
| 217 |
246993_at |
AT5G67450
|
AZF1, zinc-finger protein 1, ZF1, zinc-finger protein 1 |
0.518 |
2.523 |
| 218 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.517 |
3.189 |
| 219 |
248276_at |
AT5G53550
|
ATYSL3, YELLOW STRIPE LIKE 3, YSL3, YELLOW STRIPE like 3 |
0.516 |
1.638 |
| 220 |
249580_at |
AT5G37740
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.514 |
2.785 |
| 221 |
247765_at |
AT5G58860
|
CYP86, CYP86A1, cytochrome P450, family 86, subfamily A, polypeptide 1, HORST, hydroxylase of root suberized tissue |
0.513 |
0.065 |
| 222 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
0.513 |
5.081 |
| 223 |
260060_at |
AT1G73680
|
ALPHA DOX2, alpha dioxygenase, alpha-DOX2, alpha-dioxygenase 2 |
0.512 |
-0.376 |
| 224 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.511 |
4.305 |
| 225 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
0.510 |
3.736 |
| 226 |
266017_at |
AT2G18690
|
unknown |
0.510 |
7.480 |
| 227 |
251790_at |
AT3G55470
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.510 |
3.858 |
| 228 |
261023_at |
AT1G12200
|
FMO, flavin monooxygenase |
0.509 |
2.959 |
| 229 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.509 |
2.786 |
| 230 |
250480_at |
AT5G10290
|
[leucine-rich repeat transmembrane protein kinase family protein] |
0.508 |
0.644 |
| 231 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
0.507 |
5.619 |
| 232 |
264746_at |
AT1G62300
|
ATWRKY6, WRKY6 |
0.507 |
5.850 |
| 233 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.507 |
-0.735 |
| 234 |
252882_at |
AT4G39675
|
unknown |
0.506 |
1.990 |
| 235 |
253392_at |
AT4G32650
|
ATKC1, ARABIDOPSIS THALIANA K+ RECTIFYING CHANNEL 1, AtLKT1, A. thaliana low-K+-tolerant 1, KAT3, potassium channel in Arabidopsis thaliana 3, KC1 |
0.505 |
0.018 |
| 236 |
259735_at |
AT1G64405
|
unknown |
0.504 |
0.320 |
| 237 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
0.504 |
3.623 |
| 238 |
255342_at |
AT4G04510
|
CRK38, cysteine-rich RLK (RECEPTOR-like protein kinase) 38 |
0.504 |
5.673 |
| 239 |
245305_at |
AT4G17215
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.504 |
0.387 |
| 240 |
266658_at |
AT2G25735
|
unknown |
0.501 |
6.262 |
| 241 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
0.499 |
-0.664 |
| 242 |
263565_at |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
0.499 |
5.721 |
| 243 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
0.498 |
0.225 |
| 244 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.498 |
2.944 |
| 245 |
258566_at |
AT3G04110
|
ATGLR1.1, GLR1, GLUTAMATE RECEPTOR 1, GLR1.1, glutamate receptor 1.1 |
0.497 |
1.285 |
| 246 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.495 |
5.507 |
| 247 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
0.492 |
6.089 |
| 248 |
265471_at |
AT2G37130
|
[Peroxidase superfamily protein] |
0.491 |
0.461 |
| 249 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
0.490 |
1.039 |
| 250 |
253626_at |
AT4G30640
|
[RNI-like superfamily protein] |
0.487 |
1.279 |
| 251 |
251200_at |
AT3G63010
|
ATGID1B, GID1B, GA INSENSITIVE DWARF1B |
0.486 |
2.834 |
| 252 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.486 |
3.227 |
| 253 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
0.486 |
2.754 |
| 254 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
0.485 |
-1.206 |
| 255 |
245401_at |
AT4G17670
|
unknown |
0.484 |
2.070 |
| 256 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
0.483 |
7.334 |
| 257 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.483 |
7.743 |
| 258 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.483 |
5.192 |
| 259 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
0.483 |
5.882 |
| 260 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.482 |
5.431 |
| 261 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
0.481 |
-1.458 |
| 262 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.481 |
5.748 |
| 263 |
247819_at |
AT5G58350
|
WNK4, with no lysine (K) kinase 4, ZIK2 |
0.480 |
0.913 |
| 264 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
0.478 |
1.698 |
| 265 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.476 |
2.311 |
| 266 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.472 |
4.969 |
| 267 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.472 |
4.812 |
| 268 |
252862_at |
AT4G39830
|
[Cupredoxin superfamily protein] |
0.470 |
4.137 |
| 269 |
254453_at |
AT4G21120
|
AAT1, amino acid transporter 1, CAT1, CATIONIC AMINO ACID TRANSPORTER 1 |
0.469 |
4.530 |
| 270 |
264774_at |
AT1G22890
|
unknown |
0.469 |
4.693 |
| 271 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.467 |
3.669 |
| 272 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.467 |
3.619 |
| 273 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.466 |
0.967 |
| 274 |
265028_at |
AT1G24530
|
[Transducin/WD40 repeat-like superfamily protein] |
0.465 |
2.675 |
| 275 |
249346_at |
AT5G40780
|
LHT1, lysine histidine transporter 1 |
0.463 |
3.361 |
| 276 |
248037_at |
AT5G55930
|
ATOPT1, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 1, OPT1, oligopeptide transporter 1 |
0.463 |
2.577 |
| 277 |
260396_at |
AT1G69720
|
HO3, heme oxygenase 3 |
0.461 |
-0.087 |
| 278 |
256192_at |
AT1G30110
|
ATNUDX25, nudix hydrolase homolog 25, NUDX25, nudix hydrolase homolog 25 |
0.459 |
-0.281 |
| 279 |
256170_at |
AT1G51790
|
[Leucine-rich repeat protein kinase family protein] |
0.459 |
3.229 |
| 280 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.456 |
3.474 |
| 281 |
246968_at |
AT5G24870
|
[RING/U-box superfamily protein] |
0.456 |
-0.982 |
| 282 |
264767_at |
AT1G61380
|
SD1-29, S-domain-1 29 |
0.456 |
2.998 |
| 283 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.455 |
4.107 |
| 284 |
260623_at |
AT1G08090
|
ACH1, ATNRT2.1, NITRATE TRANSPORTER 2.1, ATNRT2:1, nitrate transporter 2:1, LIN1, LATERAL ROOT INITIATION 1, NRT2, NITRATE TRANSPORTER 2, NRT2.1, NITRATE TRANSPORTER 2.1, NRT2:1, nitrate transporter 2:1, NRT2;1AT |
0.455 |
-1.137 |
| 285 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.453 |
2.147 |
| 286 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
0.452 |
4.472 |
| 287 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.450 |
4.923 |
| 288 |
246125_at |
AT5G19875
|
unknown |
0.449 |
-0.784 |
| 289 |
252572_at |
AT3G45290
|
ATMLO3, MILDEW RESISTANCE LOCUS O 3, MLO3, MILDEW RESISTANCE LOCUS O 3 |
0.448 |
1.931 |
| 290 |
260648_at |
AT1G08050
|
[Zinc finger (C3HC4-type RING finger) family protein] |
0.447 |
4.599 |
| 291 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
0.447 |
-0.172 |
| 292 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.447 |
1.047 |
| 293 |
255527_at |
AT4G02360
|
unknown |
0.447 |
1.519 |
| 294 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.444 |
3.734 |
| 295 |
264751_at |
AT1G23020
|
ATFRO3, FERRIC REDUCTION OXIDASE 3, FRO3, ferric reduction oxidase 3 |
0.443 |
-0.020 |
| 296 |
251739_at |
AT3G56170
|
CAN, Ca-2+ dependent nuclease, CAN1, calcium dependent nuclease 1 |
0.441 |
1.475 |
| 297 |
263998_at |
AT2G22510
|
[hydroxyproline-rich glycoprotein family protein] |
0.440 |
-0.524 |
| 298 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
0.438 |
0.277 |
| 299 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
0.437 |
0.517 |
| 300 |
252309_at |
AT3G49340
|
[Cysteine proteinases superfamily protein] |
0.436 |
1.506 |
| 301 |
256096_at |
AT1G13650
|
unknown |
-0.810 |
-4.163 |
| 302 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.724 |
0.442 |
| 303 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.715 |
1.135 |
| 304 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.639 |
-4.674 |
| 305 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.636 |
1.633 |
| 306 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.612 |
0.680 |
| 307 |
245747_at |
AT1G51100
|
CRR41, CHLORORESPIRATORY REDUCTION 41 |
-0.583 |
-1.522 |
| 308 |
251788_at |
AT3G55420
|
unknown |
-0.537 |
-1.129 |
| 309 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
-0.523 |
-1.645 |
| 310 |
251665_at |
AT3G57040
|
ARR9, response regulator 9, ATRR4, RESPONSE REGULATOR 4 |
-0.521 |
-1.979 |
| 311 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
-0.517 |
-0.383 |
| 312 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.510 |
-1.106 |
| 313 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.502 |
-3.698 |
| 314 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-0.488 |
0.188 |
| 315 |
249932_at |
AT5G22390
|
unknown |
-0.480 |
-1.584 |
| 316 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.479 |
-2.068 |
| 317 |
260112_at |
AT1G63310
|
unknown |
-0.476 |
0.293 |
| 318 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.471 |
-1.192 |
| 319 |
253424_at |
AT4G32330
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.471 |
-1.341 |
| 320 |
263597_at |
AT2G01870
|
unknown |
-0.462 |
-2.429 |
| 321 |
250167_at |
AT5G15310
|
ATMIXTA, ATMYB16, myb domain protein 16, MYB16, myb domain protein 16 |
-0.458 |
-2.933 |
| 322 |
259001_at |
AT3G01960
|
unknown |
-0.456 |
-1.432 |
| 323 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
-0.445 |
0.718 |
| 324 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.441 |
-0.429 |
| 325 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.440 |
-0.308 |
| 326 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.432 |
-3.095 |
| 327 |
249939_at |
AT5G22430
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.426 |
0.225 |
| 328 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.425 |
-3.125 |
| 329 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.425 |
-2.774 |
| 330 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.420 |
-3.990 |
| 331 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.420 |
-2.341 |
| 332 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
-0.418 |
-2.008 |
| 333 |
245743_at |
AT1G51080
|
unknown |
-0.415 |
-2.425 |
| 334 |
258468_at |
AT3G06070
|
unknown |
-0.414 |
-0.618 |
| 335 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
-0.413 |
-1.292 |
| 336 |
252073_at |
AT3G51750
|
unknown |
-0.412 |
-1.757 |
| 337 |
252034_at |
AT3G52040
|
unknown |
-0.410 |
0.169 |
| 338 |
257714_at |
AT3G27360
|
H3.1, histone 3.1 |
-0.409 |
-0.141 |
| 339 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
-0.407 |
-0.034 |
| 340 |
263829_at |
AT2G40435
|
unknown |
-0.404 |
-2.826 |
| 341 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
-0.403 |
-0.857 |
| 342 |
258333_at |
AT3G16000
|
MFP1, MAR binding filament-like protein 1 |
-0.403 |
-1.407 |
| 343 |
259642_at |
AT1G69030
|
[BSD domain-containing protein] |
-0.400 |
1.537 |
| 344 |
265441_at |
AT2G20870
|
[cell wall protein precursor, putative] |
-0.397 |
-0.012 |
| 345 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
-0.394 |
-2.593 |
| 346 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
-0.394 |
-0.896 |
| 347 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.393 |
-1.815 |
| 348 |
267637_at |
AT2G42190
|
unknown |
-0.389 |
-1.932 |
| 349 |
263016_at |
AT1G23410
|
[Ribosomal protein S27a / Ubiquitin family protein] |
-0.385 |
-0.608 |
| 350 |
249916_at |
AT5G22880
|
H2B, HISTONE H2B, HTB2, histone B2 |
-0.384 |
-0.539 |
| 351 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.384 |
-2.216 |
| 352 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.383 |
0.700 |
| 353 |
253679_at |
AT4G29610
|
[Cytidine/deoxycytidylate deaminase family protein] |
-0.381 |
-3.204 |
| 354 |
245426_at |
AT4G17540
|
unknown |
-0.381 |
-1.057 |
| 355 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.381 |
0.415 |
| 356 |
247942_at |
AT5G57120
|
unknown |
-0.380 |
-0.209 |
| 357 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.380 |
-3.989 |
| 358 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.378 |
-2.189 |
| 359 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.377 |
-3.494 |
| 360 |
259658_at |
AT1G55370
|
NDF5, NDH-dependent cyclic electron flow 5 |
-0.375 |
-1.869 |
| 361 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.374 |
-2.050 |
| 362 |
260012_at |
AT1G67865
|
unknown |
-0.370 |
0.332 |
| 363 |
267509_at |
AT2G45660
|
AGL20, AGAMOUS-like 20, ATSOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1, SOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1 |
-0.368 |
-1.689 |
| 364 |
248177_at |
AT5G54630
|
[zinc finger protein-related] |
-0.368 |
-2.316 |
| 365 |
247955_at |
AT5G56950
|
NAP1;3, nucleosome assembly protein 1;3, NFA03, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP A 03, NFA3, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP A3 |
-0.367 |
0.967 |
| 366 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
-0.367 |
1.049 |
| 367 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.364 |
-2.084 |
| 368 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.361 |
1.982 |
| 369 |
259373_at |
AT1G69160
|
unknown |
-0.360 |
-1.405 |
| 370 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.359 |
-1.096 |
| 371 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.358 |
-0.132 |
| 372 |
248975_at |
AT5G45040
|
CYTC6A, cytochrome c6A |
-0.357 |
-0.728 |
| 373 |
248537_at |
AT5G50100
|
[Putative thiol-disulphide oxidoreductase DCC] |
-0.357 |
-0.705 |
| 374 |
262039_at |
AT1G80050
|
APT2, adenine phosphoribosyl transferase 2, ATAPT2, PHT1.1 |
-0.357 |
-1.062 |
| 375 |
255028_at |
AT4G09890
|
unknown |
-0.353 |
-2.284 |
| 376 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.352 |
-2.862 |
| 377 |
255793_at |
AT2G33250
|
unknown |
-0.352 |
-1.230 |
| 378 |
249410_at |
AT5G40380
|
CRK42, cysteine-rich RLK (RECEPTOR-like protein kinase) 42 |
-0.351 |
-2.852 |
| 379 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
-0.350 |
-1.924 |
| 380 |
246792_at |
AT5G27290
|
unknown |
-0.350 |
-0.893 |
| 381 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
-0.349 |
-0.081 |
| 382 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.348 |
-3.894 |
| 383 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.348 |
-2.763 |
| 384 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.347 |
-0.561 |
| 385 |
265819_at |
AT2G17972
|
unknown |
-0.346 |
-1.377 |
| 386 |
266423_at |
AT2G41340
|
RPB5D, RNA polymerase II fifth largest subunit, D |
-0.343 |
0.193 |
| 387 |
248623_at |
AT5G49170
|
unknown |
-0.342 |
0.738 |
| 388 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
-0.342 |
-3.209 |
| 389 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
-0.342 |
-0.602 |
| 390 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.341 |
-3.475 |
| 391 |
245088_at |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
-0.340 |
-0.741 |
| 392 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.339 |
-3.276 |
| 393 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.336 |
-1.121 |
| 394 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.335 |
-3.560 |
| 395 |
250031_at |
AT5G18240
|
ATMYR1, ARABIDOPSIS MYB-RELATED PROTEIN 1, MYR1, myb-related protein 1 |
-0.333 |
-0.331 |
| 396 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.331 |
-2.402 |
| 397 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.330 |
-0.903 |
| 398 |
253251_at |
AT4G34730
|
RBF1, RbfA domain-containing protein 1 |
-0.328 |
-0.940 |
| 399 |
254683_at |
AT4G13800
|
unknown |
-0.327 |
-0.249 |
| 400 |
256599_at |
AT3G14760
|
unknown |
-0.324 |
-1.445 |
| 401 |
260041_at |
AT1G68780
|
[RNI-like superfamily protein] |
-0.324 |
-1.695 |
| 402 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.324 |
-2.721 |
| 403 |
256455_at |
AT1G75190
|
unknown |
-0.323 |
-1.144 |
| 404 |
267057_at |
AT2G32500
|
[Stress responsive alpha-beta barrel domain protein] |
-0.323 |
-1.013 |
| 405 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.322 |
-1.581 |
| 406 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.322 |
-0.758 |
| 407 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
-0.321 |
-1.808 |
| 408 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.321 |
-3.716 |
| 409 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
-0.319 |
-2.792 |
| 410 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
-0.318 |
-1.881 |
| 411 |
246231_at |
AT4G37080
|
unknown |
-0.318 |
-2.238 |
| 412 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.317 |
-2.904 |
| 413 |
260125_at |
AT1G36390
|
[Co-chaperone GrpE family protein] |
-0.316 |
-0.672 |
| 414 |
261772_at |
AT1G76240
|
unknown |
-0.316 |
-1.787 |
| 415 |
262120_at |
AT1G02950
|
ATGSTF4, glutathione S-transferase F4, GST31, GLUTATHIONE S-TRANSFERASE 31, GSTF4, glutathione S-transferase F4 |
-0.315 |
0.262 |
| 416 |
261502_at |
AT1G14440
|
AtHB31, homeobox protein 31, FTM2, FLORAL TRANSITION AT THE MERISTEM2, HB31, homeobox protein 31, ZHD4, ZINC FINGER HOMEODOMAIN 4 |
-0.314 |
-0.630 |
| 417 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.314 |
-3.398 |
| 418 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.314 |
-2.439 |
| 419 |
246411_at |
AT1G57770
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.314 |
-0.887 |
| 420 |
252023_at |
AT3G52920
|
unknown |
-0.313 |
-1.468 |
| 421 |
259230_at |
AT3G07780
|
OBE1, OBERON1 |
-0.313 |
2.469 |
| 422 |
248247_at |
AT5G53210
|
SPCH, SPEECHLESS |
-0.312 |
0.017 |
| 423 |
263251_at |
AT2G31410
|
unknown |
-0.311 |
0.306 |
| 424 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.311 |
0.400 |
| 425 |
254794_at |
AT4G12970
|
EPFL9, STOMAGEN, STOMAGEN |
-0.311 |
-1.572 |
| 426 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
-0.311 |
-0.582 |
| 427 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
-0.310 |
-1.662 |
| 428 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.310 |
-2.593 |
| 429 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
-0.310 |
-2.151 |
| 430 |
246103_at |
AT5G28640
|
AN3, ANGUSTIFOLIA 3, ATGIF1, ARABIDOPSIS GRF1-INTERACTING FACTOR 1, GIF, GRF1-INTERACTING FACTOR, GIF1, GRF1-INTERACTING FACTOR 1 |
-0.308 |
0.097 |
| 431 |
245353_at |
AT4G16000
|
unknown |
-0.308 |
1.358 |
| 432 |
266384_at |
AT2G14660
|
unknown |
-0.308 |
-1.792 |
| 433 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
-0.307 |
-2.829 |
| 434 |
256675_at |
AT3G52170
|
[DNA binding] |
-0.306 |
-0.751 |
| 435 |
259586_at |
AT1G28100
|
unknown |
-0.306 |
-0.801 |
| 436 |
250179_at |
AT5G14440
|
[Surfeit locus protein 2 (SURF2)] |
-0.304 |
-0.064 |
| 437 |
250762_at |
AT5G05990
|
[Mitochondrial glycoprotein family protein] |
-0.303 |
-1.014 |
| 438 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-0.303 |
-1.660 |
| 439 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.302 |
-1.897 |
| 440 |
260076_at |
AT1G73630
|
[EF hand calcium-binding protein family] |
-0.301 |
-1.148 |
| 441 |
266489_at |
AT2G35190
|
ATNPSN11, NPSN11, novel plant snare 11, NSPN11 |
-0.300 |
0.426 |
| 442 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
-0.300 |
-1.827 |
| 443 |
267580_at |
AT2G41990
|
unknown |
-0.299 |
-0.895 |
| 444 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
-0.298 |
-0.715 |
| 445 |
264025_at |
AT2G21050
|
LAX2, like AUXIN RESISTANT 2 |
-0.297 |
-1.894 |
| 446 |
252025_at |
AT3G52900
|
unknown |
-0.297 |
-0.169 |
| 447 |
251141_at |
AT5G01075
|
[Glycosyl hydrolase family 35 protein] |
-0.297 |
-1.961 |
| 448 |
263264_at |
AT2G38810
|
HTA8, histone H2A 8 |
-0.296 |
0.001 |
| 449 |
257937_at |
AT3G19810
|
unknown |
-0.296 |
-0.654 |
| 450 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
-0.295 |
-1.942 |
| 451 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.295 |
-2.363 |
| 452 |
260332_at |
AT1G70470
|
unknown |
-0.295 |
-0.652 |
| 453 |
245027_at |
AT2G26550
|
HO2, heme oxygenase 2 |
-0.295 |
-0.906 |
| 454 |
250751_at |
AT5G05890
|
[UDP-Glycosyltransferase superfamily protein] |
-0.293 |
-2.087 |
| 455 |
253635_at |
AT4G30620
|
[Uncharacterised BCR, YbaB family COG0718] |
-0.293 |
-1.252 |
| 456 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.293 |
-3.188 |
| 457 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
-0.293 |
-0.997 |
| 458 |
253254_at |
AT4G34650
|
SQS2, squalene synthase 2 |
-0.292 |
-1.434 |
| 459 |
249958_at |
AT5G18970
|
[AWPM-19-like family protein] |
-0.292 |
0.052 |
| 460 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
-0.292 |
-0.225 |
| 461 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.291 |
-0.082 |
| 462 |
265874_at |
AT2G01640
|
unknown |
-0.291 |
0.477 |
| 463 |
267549_at |
AT2G32640
|
[Lycopene beta/epsilon cyclase protein] |
-0.290 |
-0.911 |
| 464 |
252147_at |
AT3G51270
|
[protein serine/threonine kinases] |
-0.288 |
0.144 |
| 465 |
255817_at |
AT2G33330
|
PDLP3, plasmodesmata-located protein 3 |
-0.288 |
-1.479 |
| 466 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-0.288 |
-2.146 |
| 467 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.287 |
-1.536 |
| 468 |
256741_at |
AT3G29375
|
[XH domain-containing protein] |
-0.286 |
-1.236 |
| 469 |
263535_at |
AT2G24970
|
unknown |
-0.285 |
-0.775 |
| 470 |
251669_at |
AT3G57180
|
BPG2, BRASSINAZOLE(BRZ) INSENSITIVE PALE GREEN 2 |
-0.285 |
-1.100 |
| 471 |
247074_at |
AT5G66590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.284 |
-3.462 |
| 472 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.284 |
-2.472 |
| 473 |
267127_at |
AT2G23610
|
ATMES3, ARABIDOPSIS THALIANA METHYL ESTERASE 3, MES3, methyl esterase 3 |
-0.283 |
-1.198 |
| 474 |
259207_at |
AT3G09050
|
unknown |
-0.283 |
-1.522 |
| 475 |
256441_at |
AT3G10940
|
LSF2, LIKE SEX4 2 |
-0.282 |
-1.515 |
| 476 |
258386_at |
AT3G15520
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.282 |
-0.283 |
| 477 |
266957_at |
AT2G34640
|
HMR, HEMERA, PTAC12, plastid transcriptionally active 12, TAC12 |
-0.281 |
-0.512 |
| 478 |
252361_at |
AT3G48490
|
unknown |
-0.281 |
-0.212 |
| 479 |
250379_at |
AT5G11590
|
TINY2, TINY2 |
-0.280 |
-3.651 |
| 480 |
264697_at |
AT1G70210
|
ATCYCD1;1, CYCD1;1, CYCLIN D1;1 |
-0.279 |
-1.471 |
| 481 |
259889_at |
AT1G76405
|
unknown |
-0.278 |
-0.576 |
| 482 |
250753_at |
AT5G05860
|
UGT76C2, UDP-glucosyl transferase 76C2 |
-0.278 |
0.574 |
| 483 |
249920_at |
AT5G19260
|
FAF3, FANTASTIC FOUR 3 |
-0.278 |
0.737 |
| 484 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.277 |
-1.507 |
| 485 |
253811_at |
AT4G28190
|
ULT, ULTRAPETALA, ULT1, ULTRAPETALA1 |
-0.277 |
-1.014 |
| 486 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.276 |
-2.285 |
| 487 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
-0.276 |
-0.700 |
| 488 |
245165_at |
AT2G33180
|
unknown |
-0.276 |
-1.162 |
| 489 |
263674_at |
AT2G04790
|
unknown |
-0.276 |
-2.575 |
| 490 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.275 |
-0.924 |
| 491 |
246303_at |
AT3G51870
|
[Mitochondrial substrate carrier family protein] |
-0.275 |
-0.723 |
| 492 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.275 |
-0.595 |
| 493 |
250394_at |
AT5G10910
|
[mraW methylase family protein] |
-0.275 |
-0.970 |
| 494 |
245291_at |
AT4G16155
|
[dihydrolipoyl dehydrogenases] |
-0.275 |
-0.549 |
| 495 |
259180_at |
AT3G01680
|
AtSEOR1, Arabidopsis thaliana sieve element occlusion-related 1, SEOb, sieve element occlusion b, SEOR1, Sieve-Element-Occlusion-Related 1 |
-0.274 |
0.210 |
| 496 |
247910_at |
AT5G57410
|
[Afadin/alpha-actinin-binding protein] |
-0.273 |
-0.040 |
| 497 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.273 |
-2.299 |
| 498 |
263515_at |
AT2G21640
|
unknown |
-0.273 |
2.167 |
| 499 |
245417_at |
AT4G17360
|
[Formyl transferase] |
-0.272 |
-1.558 |
| 500 |
246487_at |
AT5G16030
|
unknown |
-0.272 |
-2.979 |
| 501 |
265960_at |
AT2G37470
|
[Histone superfamily protein] |
-0.272 |
-0.108 |
| 502 |
267038_at |
AT2G38480
|
[Uncharacterised protein family (UPF0497)] |
-0.271 |
-1.308 |
| 503 |
259038_at |
AT3G09210
|
PTAC13, plastid transcriptionally active 13 |
-0.270 |
-1.029 |
| 504 |
266478_at |
AT2G31170
|
FIONA, FIONA, SYCO ARATH, cysteinyl t-RNA synthetase |
-0.270 |
-0.929 |
| 505 |
256978_at |
AT3G21110
|
ATPURC, PUR7, purin 7, PURC, PURIN C |
-0.270 |
-0.813 |
| 506 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
-0.269 |
-3.356 |
| 507 |
264343_at |
AT1G11850
|
unknown |
-0.269 |
-0.422 |
| 508 |
247878_at |
AT5G57760
|
unknown |
-0.269 |
4.101 |
| 509 |
251933_at |
AT3G54060
|
unknown |
-0.268 |
-1.024 |
| 510 |
247415_at |
AT5G63060
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.267 |
-1.093 |
| 511 |
257794_at |
AT3G27050
|
unknown |
-0.267 |
-1.195 |
| 512 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.267 |
-1.697 |
| 513 |
266760_at |
AT2G46870
|
NGA1, NGATHA1 |
-0.267 |
-0.579 |
| 514 |
251586_at |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
-0.267 |
-4.102 |
| 515 |
247716_at |
AT5G59350
|
unknown |
-0.266 |
-2.065 |
| 516 |
248753_at |
AT5G47630
|
mtACP3, mitochondrial acyl carrier protein 3 |
-0.266 |
-1.017 |
| 517 |
255285_at |
AT4G04630
|
unknown |
-0.266 |
0.481 |
| 518 |
263688_at |
AT1G26920
|
unknown |
-0.265 |
-2.357 |
| 519 |
247486_at |
AT5G62140
|
unknown |
-0.265 |
-3.202 |
| 520 |
263802_at |
AT2G40430
|
unknown |
-0.265 |
-0.157 |
| 521 |
260676_at |
AT1G19450
|
[Major facilitator superfamily protein] |
-0.265 |
-1.604 |
| 522 |
249060_at |
AT5G44560
|
VPS2.2 |
-0.263 |
-0.037 |
| 523 |
259981_at |
AT1G76450
|
[Photosystem II reaction center PsbP family protein] |
-0.263 |
-0.686 |
| 524 |
261861_at |
AT1G50450
|
[Saccharopine dehydrogenase ] |
-0.263 |
-0.745 |
| 525 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.262 |
-1.683 |
| 526 |
246962_s_at |
AT5G24800
|
ATBZIP9, ARABIDOPSIS THALIANA BASIC LEUCINE ZIPPER 9, BZIP9, basic leucine zipper 9, BZO2H2, BASIC LEUCINE ZIPPER O2 HOMOLOG 2 |
-0.262 |
1.236 |
| 527 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.261 |
-1.165 |
| 528 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.261 |
-2.201 |
| 529 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
-0.260 |
-3.247 |
| 530 |
249900_at |
AT5G22640
|
emb1211, embryo defective 1211, TIC100, TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 100 |
-0.260 |
-1.470 |
| 531 |
266521_at |
AT2G24020
|
[Uncharacterised BCR, YbaB family COG0718] |
-0.259 |
-0.135 |
| 532 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.259 |
-0.286 |
| 533 |
246936_at |
AT5G25360
|
unknown |
-0.258 |
0.176 |
| 534 |
252574_at |
AT3G45430
|
LecRK-I.5, L-type lectin receptor kinase I.5 |
-0.258 |
1.676 |
| 535 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.257 |
-2.384 |
| 536 |
249886_at |
AT5G22320
|
[Leucine-rich repeat (LRR) family protein] |
-0.257 |
0.200 |
| 537 |
247903_at |
AT5G57340
|
unknown |
-0.257 |
-1.123 |
| 538 |
266402_at |
AT2G38780
|
unknown |
-0.257 |
-0.750 |
| 539 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
-0.256 |
-1.697 |
| 540 |
266226_at |
AT2G28740
|
HIS4, histone H4 |
-0.256 |
-0.263 |
| 541 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-0.255 |
-0.236 |
| 542 |
255362_at |
AT4G04000
|
[non-LTR retrotransposon family (LINE), has a 2.6e-39 P-value blast match to GB:AAB41224 ORF2 (LINE-element) (Rattus norvegicus)] |
-0.255 |
0.225 |
| 543 |
250327_at |
AT5G12050
|
unknown |
-0.255 |
-1.784 |
| 544 |
256066_at |
AT1G06980
|
unknown |
-0.255 |
-1.283 |
| 545 |
248493_at |
AT5G51100
|
FSD2, Fe superoxide dismutase 2 |
-0.254 |
-0.059 |
| 546 |
255779_at |
AT1G18650
|
PDCB3, plasmodesmata callose-binding protein 3 |
-0.253 |
-1.460 |
| 547 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
-0.253 |
-0.777 |
| 548 |
260308_at |
AT1G70610
|
ABCB26, ATP-binding cassette B26, ATTAP1, transporter associated with antigen processing protein 1, TAP1, transporter associated with antigen processing protein 1 |
-0.253 |
-0.689 |
| 549 |
264078_at |
AT2G28470
|
BGAL8, beta-galactosidase 8 |
-0.252 |
-2.094 |
| 550 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.252 |
-1.448 |
| 551 |
258796_at |
AT3G04630
|
WDL1, WVD2-like 1 |
-0.252 |
0.606 |
| 552 |
261256_at |
AT1G05760
|
RTM1, restricted tev movement 1 |
-0.251 |
0.225 |
| 553 |
247733_at |
AT5G59610
|
DJC73, DNA J protein C73 |
-0.250 |
-0.565 |
| 554 |
266253_at |
AT2G27840
|
HD2D, histone deacetylase 2D, HDA13, HISTONE DEACETYLASE 13, HDT04, HDT4 |
-0.250 |
-0.757 |
| 555 |
263840_at |
AT2G36885
|
unknown |
-0.250 |
-0.647 |
| 556 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
-0.250 |
-1.588 |
| 557 |
252227_at |
AT3G49900
|
[Phototropic-responsive NPH3 family protein] |
-0.250 |
-1.156 |
| 558 |
251768_at |
AT3G55940
|
[Phosphoinositide-specific phospholipase C family protein] |
-0.250 |
-1.744 |
| 559 |
246265_at |
AT1G31860
|
AT-IE, HISN2, HISTIDINE BIOSYNTHESIS 2 |
-0.248 |
-0.440 |
| 560 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.248 |
-2.583 |
| 561 |
261406_at |
AT1G18800
|
NRP2, NAP1-related protein 2 |
-0.248 |
-0.135 |
| 562 |
255053_at |
AT4G09730
|
RH39, RH39 |
-0.248 |
-0.908 |
| 563 |
247214_at |
AT5G64850
|
unknown |
-0.248 |
-1.362 |
| 564 |
259801_at |
AT1G72230
|
[Cupredoxin superfamily protein] |
-0.247 |
-1.149 |
| 565 |
266481_at |
AT2G31070
|
TCP10, TCP domain protein 10 |
-0.247 |
-1.406 |
| 566 |
259160_at |
AT3G05410
|
[Photosystem II reaction center PsbP family protein] |
-0.247 |
-1.905 |
| 567 |
254427_at |
AT4G21190
|
emb1417, embryo defective 1417 |
-0.247 |
-1.591 |
| 568 |
260685_at |
AT1G17650
|
AtGLYR2, GLYR2, glyoxylate reductase 2, GR2, GLYOXYLATE REDUCTASE 2 |
-0.247 |
-0.387 |
| 569 |
251463_at |
AT3G59800
|
unknown |
-0.246 |
-0.104 |
| 570 |
265656_at |
AT2G13820
|
AtXYP2, XYP2, xylogen protein 2 |
-0.246 |
-0.939 |
| 571 |
249927_at |
AT5G19220
|
ADG2, ADP GLUCOSE PYROPHOSPHORYLASE 2, APL1, ADP glucose pyrophosphorylase large subunit 1 |
-0.246 |
-0.474 |
| 572 |
263763_at |
AT2G21385
|
unknown |
-0.245 |
-0.221 |
| 573 |
267260_at |
AT2G23130
|
AGP17, arabinogalactan protein 17, ATAGP17 |
-0.245 |
-2.886 |
| 574 |
247899_at |
AT5G57345
|
unknown |
-0.245 |
-1.611 |
| 575 |
254612_at |
AT4G19100
|
PAM68, photosynthesis affected mutant 68 |
-0.244 |
-0.862 |
| 576 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.244 |
1.879 |
| 577 |
258884_at |
AT3G10050
|
OMR1, L-O-methylthreonine resistant 1 |
-0.244 |
0.164 |
| 578 |
266916_at |
AT2G45860
|
unknown |
-0.244 |
-0.316 |
| 579 |
248094_at |
AT5G55220
|
[trigger factor type chaperone family protein] |
-0.243 |
-0.760 |
| 580 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.243 |
-3.179 |
| 581 |
254911_at |
AT4G11100
|
unknown |
-0.243 |
-0.796 |
| 582 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.243 |
-3.608 |
| 583 |
256065_at |
AT1G07070
|
[Ribosomal protein L35Ae family protein] |
-0.242 |
-0.699 |
| 584 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.242 |
-1.000 |
| 585 |
252366_at |
AT3G48420
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.242 |
-0.487 |
| 586 |
266942_at |
AT2G18990
|
TXND9, thioredoxin domain-containing protein 9 homolog |
-0.241 |
-0.226 |
| 587 |
261058_at |
AT1G01370
|
CENH3, CENTROMERIC HISTONE H3, HTR12 |
-0.241 |
0.305 |
| 588 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.240 |
-1.783 |
| 589 |
260982_at |
AT1G53520
|
AtFAP3, FAP3, fatty-acid-binding protein 3 |
-0.240 |
-0.343 |
| 590 |
250980_at |
AT5G03130
|
unknown |
-0.240 |
-2.133 |
| 591 |
248924_at |
AT5G45960
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.240 |
-0.882 |
| 592 |
253537_at |
AT4G31560
|
HCF153, high chlorophyll fluorescence 153 |
-0.240 |
-1.092 |
| 593 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.240 |
-1.868 |
| 594 |
248709_at |
AT5G48470
|
PRDA1, PEP-Related Development Arrested 1 |
-0.238 |
-0.398 |
| 595 |
265704_at |
AT2G03420
|
unknown |
-0.238 |
0.225 |
| 596 |
246196_at |
AT4G37090
|
unknown |
-0.237 |
0.191 |
| 597 |
260872_at |
AT1G21350
|
[Thioredoxin superfamily protein] |
-0.236 |
-0.560 |
| 598 |
262288_at |
AT1G70760
|
CRR23, CHLORORESPIRATORY REDUCTION 23, NdhL, NADH dehydrogenase-like complex L |
-0.236 |
-1.319 |
| 599 |
257825_at |
AT3G26700
|
[Protein kinase superfamily protein] |
-0.236 |
-1.510 |
| 600 |
264781_at |
AT1G08540
|
ABC1, ATSIG1, SIGMA FACTOR 1, ATSIG2, SIGMA FACTOR 2, SIG1, RNA POLYMERASE SIGMA SUBUNIT 1, SIG2, RNApolymerase sigma subunit 2, SIGA, SIGB, SIGMA FACTOR B |
-0.236 |
-0.993 |