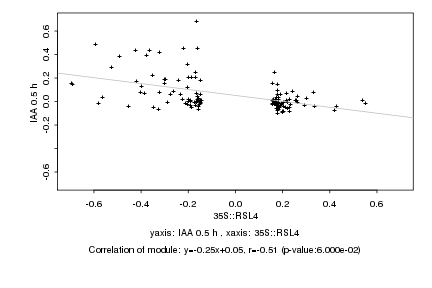
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
35S::RSL4 |
Log2 signal ratio
IAA 0.5 h |
| 1 |
261647_at |
AT1G27740
|
RSL4, root hair defective 6-like 4 |
0.549 |
-0.015 |
| 2 |
249686_at |
AT5G36140
|
CYP716A2, cytochrome P450, family 716, subfamily A, polypeptide 2 |
0.535 |
0.016 |
| 3 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.425 |
-0.037 |
| 4 |
249102_at |
AT5G43590
|
[Acyl transferase/acyl hydrolase/lysophospholipase superfamily protein] |
0.419 |
-0.076 |
| 5 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.334 |
-0.038 |
| 6 |
245036_at |
AT2G26410
|
Iqd4, IQ-domain 4 |
0.329 |
0.083 |
| 7 |
266674_at |
AT2G29620
|
unknown |
0.297 |
0.026 |
| 8 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
0.289 |
-0.029 |
| 9 |
254107_at |
AT4G25220
|
AtG3Pp2, glycerol-3-phosphate permease 2, G3Pp2, glycerol-3-phosphate permease 2, RHS15, root hair specific 15 |
0.261 |
-0.002 |
| 10 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
0.260 |
0.045 |
| 11 |
259720_at |
AT1G61080
|
[Hydroxyproline-rich glycoprotein family protein] |
0.258 |
0.017 |
| 12 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
0.254 |
0.009 |
| 13 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.240 |
0.087 |
| 14 |
253353_at |
AT4G33730
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.231 |
-0.020 |
| 15 |
247781_at |
AT5G58784
|
[Undecaprenyl pyrophosphate synthetase family protein] |
0.229 |
-0.079 |
| 16 |
249773_at |
AT5G24140
|
SQP2, squalene monooxygenase 2 |
0.228 |
-0.049 |
| 17 |
254347_at |
AT4G22070
|
ATWRKY31, WRKY31, WRKY DNA-binding protein 31 |
0.226 |
0.025 |
| 18 |
250744_at |
AT5G05840
|
unknown |
0.220 |
-0.055 |
| 19 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
0.218 |
-0.006 |
| 20 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
0.218 |
0.000 |
| 21 |
257049_at |
AT3G15250
|
unknown |
0.215 |
0.071 |
| 22 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
0.215 |
0.011 |
| 23 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
0.211 |
-0.043 |
| 24 |
252872_at |
AT4G40010
|
SNRK2-7, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-7, SNRK2.7, SNF1-related protein kinase 2.7, SRK2F |
0.203 |
-0.078 |
| 25 |
264832_at |
AT1G03660
|
[Ankyrin-repeat containing protein] |
0.201 |
-0.042 |
| 26 |
254025_at |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.201 |
-0.008 |
| 27 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
0.199 |
-0.093 |
| 28 |
262865_at |
AT1G64930
|
CYP89A7, cytochrome P450, family 87, subfamily A, polypeptide 7 |
0.198 |
-0.079 |
| 29 |
247229_at |
AT5G65160
|
TPR14, tetratricopeptide repeat 14 |
0.193 |
-0.017 |
| 30 |
253910_at |
AT4G27290
|
[S-locus lectin protein kinase family protein] |
0.191 |
-0.010 |
| 31 |
247531_at |
AT5G61550
|
[U-box domain-containing protein kinase family protein] |
0.189 |
-0.032 |
| 32 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.189 |
0.063 |
| 33 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
0.186 |
-0.045 |
| 34 |
245245_at |
AT1G44318
|
hemb2 |
0.181 |
-0.060 |
| 35 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.181 |
0.018 |
| 36 |
251577_at |
AT3G58350
|
RTM3, RESTRICTED TEV MOVEMENT 3 |
0.181 |
-0.004 |
| 37 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.181 |
0.036 |
| 38 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.180 |
-0.027 |
| 39 |
264391_at |
AT1G11920
|
[Pectin lyase-like superfamily protein] |
0.180 |
-0.041 |
| 40 |
258182_at |
AT3G21500
|
DXL1, DXS-like 1, DXPS1, 1-deoxy-D-xylulose 5-phosphate synthase 1, DXS2, 1-deoxy-D-xylulose 5-phosphate (DXP) synthase 2 |
0.179 |
-0.016 |
| 41 |
246888_at |
AT5G26270
|
unknown |
0.179 |
-0.043 |
| 42 |
267128_at |
AT2G23620
|
ATMES1, ARABIDOPSIS THALIANA METHYL ESTERASE 1, MES1, methyl esterase 1 |
0.179 |
-0.014 |
| 43 |
264940_at |
AT1G60470
|
AtGolS4, galactinol synthase 4, GolS4, galactinol synthase 4 |
0.179 |
-0.012 |
| 44 |
253734_at |
AT4G29180
|
RHS16, root hair specific 16 |
0.178 |
-0.051 |
| 45 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.177 |
-0.095 |
| 46 |
251096_at |
AT5G01550
|
LecRK-VI.3, L-type lectin receptor kinase VI.3, LECRKA4.2, lectin receptor kinase a4.1 |
0.177 |
0.098 |
| 47 |
255648_at |
AT4G00910
|
[Aluminium activated malate transporter family protein] |
0.176 |
-0.071 |
| 48 |
254850_at |
AT4G12000
|
[SNARE associated Golgi protein family] |
0.176 |
-0.010 |
| 49 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.175 |
0.152 |
| 50 |
252494_at |
AT3G46760
|
LecRK-S.3, L-type lectin receptor kinase S.3 |
0.175 |
0.060 |
| 51 |
251405_at |
AT3G60330
|
AHA7, H(+)-ATPase 7, HA7, H(+)-ATPase 7 |
0.173 |
-0.027 |
| 52 |
247617_at |
AT5G60270
|
LecRK-I.7, L-type lectin receptor kinase I.7 |
0.172 |
0.000 |
| 53 |
259576_at |
AT1G35330
|
[RING/U-box superfamily protein] |
0.172 |
-0.059 |
| 54 |
250037_at |
AT5G18350
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.171 |
0.038 |
| 55 |
255620_at |
AT4G01380
|
[plastocyanin-like domain-containing protein] |
0.169 |
0.004 |
| 56 |
251466_at |
AT3G59340
|
unknown |
0.169 |
-0.024 |
| 57 |
260948_at |
AT1G06100
|
[Fatty acid desaturase family protein] |
0.168 |
0.021 |
| 58 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.165 |
-0.025 |
| 59 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.163 |
0.251 |
| 60 |
254258_at |
AT4G23410
|
TET5, tetraspanin5 |
0.163 |
-0.011 |
| 61 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
0.162 |
-0.010 |
| 62 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.158 |
0.025 |
| 63 |
260201_at |
AT1G67600
|
[Acid phosphatase/vanadium-dependent haloperoxidase-related protein] |
0.158 |
0.003 |
| 64 |
258907_at |
AT3G06370
|
ATNHX4, NHX4, sodium hydrogen exchanger 4 |
0.156 |
0.156 |
| 65 |
254562_at |
AT4G19230
|
CYP707A1, cytochrome P450, family 707, subfamily A, polypeptide 1 |
0.156 |
-0.019 |
| 66 |
265582_at |
AT2G20030
|
[RING/U-box superfamily protein] |
0.155 |
0.010 |
| 67 |
250632_at |
AT5G07450
|
CYCP4;3, cyclin p4;3 |
0.154 |
-0.010 |
| 68 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
-0.698 |
0.159 |
| 69 |
253259_at |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
-0.693 |
0.153 |
| 70 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
-0.596 |
0.488 |
| 71 |
262211_at |
AT1G74930
|
ORA47 |
-0.582 |
-0.009 |
| 72 |
248684_at |
AT5G48485
|
DIR1, DEFECTIVE IN INDUCED RESISTANCE 1 |
-0.567 |
0.040 |
| 73 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
-0.527 |
0.292 |
| 74 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
-0.494 |
0.386 |
| 75 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
-0.456 |
-0.037 |
| 76 |
253643_at |
AT4G29780
|
unknown |
-0.427 |
0.437 |
| 77 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
-0.421 |
0.176 |
| 78 |
262373_at |
AT1G73120
|
unknown |
-0.404 |
0.084 |
| 79 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
-0.401 |
0.129 |
| 80 |
250098_at |
AT5G17350
|
unknown |
-0.389 |
0.073 |
| 81 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
-0.380 |
0.398 |
| 82 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
-0.366 |
0.436 |
| 83 |
252557_at |
AT3G45960
|
ATEXLA3, expansin-like A3, ATEXPL3, ATHEXP BETA 2.3, EXLA3, expansin-like A3, EXPL3 |
-0.354 |
0.230 |
| 84 |
248822_at |
AT5G47000
|
[Peroxidase superfamily protein] |
-0.350 |
-0.049 |
| 85 |
256009_at |
AT1G19210
|
[Integrase-type DNA-binding superfamily protein] |
-0.329 |
-0.067 |
| 86 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
-0.325 |
0.081 |
| 87 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
-0.324 |
0.425 |
| 88 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
-0.304 |
0.190 |
| 89 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
-0.302 |
0.155 |
| 90 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
-0.298 |
0.192 |
| 91 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
-0.292 |
-0.001 |
| 92 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.276 |
0.062 |
| 93 |
253502_at |
AT4G31940
|
CYP82C4, cytochrome P450, family 82, subfamily C, polypeptide 4 |
-0.264 |
0.088 |
| 94 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
-0.245 |
0.185 |
| 95 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.233 |
0.065 |
| 96 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
-0.226 |
0.018 |
| 97 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
-0.223 |
0.454 |
| 98 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
-0.214 |
-0.014 |
| 99 |
251229_at |
AT3G62740
|
BGLU7, beta glucosidase 7 |
-0.207 |
-0.022 |
| 100 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
-0.207 |
0.120 |
| 101 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
-0.205 |
0.321 |
| 102 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
-0.203 |
0.022 |
| 103 |
264837_at |
AT1G03600
|
PSB27 |
-0.201 |
0.008 |
| 104 |
248186_at |
AT5G53880
|
unknown |
-0.201 |
0.004 |
| 105 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
-0.199 |
0.210 |
| 106 |
264185_at |
AT1G54780
|
AtTLP18.3, TLP18.3, thylakoid lumen protein 18.3 |
-0.193 |
0.008 |
| 107 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
-0.192 |
0.011 |
| 108 |
247358_at |
AT5G63580
|
ATFLS2, FLS2, flavonol synthase 2 |
-0.191 |
-0.029 |
| 109 |
255422_at |
AT4G03270
|
CYCD6;1, Cyclin D6;1 |
-0.189 |
-0.046 |
| 110 |
261470_at |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
-0.187 |
0.209 |
| 111 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
-0.175 |
-0.000 |
| 112 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
-0.173 |
0.255 |
| 113 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-0.173 |
0.206 |
| 114 |
265349_at |
AT2G22610
|
[Di-glucose binding protein with Kinesin motor domain] |
-0.171 |
-0.032 |
| 115 |
252073_at |
AT3G51750
|
unknown |
-0.169 |
0.073 |
| 116 |
248886_at |
AT5G46110
|
APE2, ACCLIMATION OF PHOTOSYNTHESIS TO ENVIRONMENT 2, TPT, triose-phosphate ⁄ phosphate translocator |
-0.166 |
0.000 |
| 117 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
-0.166 |
0.688 |
| 118 |
261638_at |
AT1G49975
|
[INVOLVED IN: photosynthesis] |
-0.166 |
0.006 |
| 119 |
245731_at |
AT1G73500
|
ATMKK9, MKK9, MAP kinase kinase 9 |
-0.164 |
0.049 |
| 120 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
-0.163 |
0.456 |
| 121 |
247659_at |
AT5G60040
|
NRPC1, nuclear RNA polymerase C1 |
-0.161 |
0.024 |
| 122 |
247882_at |
AT5G57785
|
unknown |
-0.160 |
-0.066 |
| 123 |
263232_at |
AT1G05700
|
[Leucine-rich repeat transmembrane protein kinase protein] |
-0.159 |
0.032 |
| 124 |
255572_at |
AT4G01050
|
TROL, thylakoid rhodanese-like |
-0.159 |
0.009 |
| 125 |
260353_at |
AT1G69230
|
SP1L2, SPIRAL1-like2 |
-0.157 |
-0.037 |
| 126 |
262970_at |
AT1G75690
|
LQY1, LOW QUANTUM YIELD OF PHOTOSYSTEM II 1 |
-0.156 |
0.007 |
| 127 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-0.154 |
-0.023 |
| 128 |
256569_at |
AT3G19550
|
unknown |
-0.152 |
0.064 |
| 129 |
246576_at |
AT1G31650
|
ATROPGEF14, ROPGEF14, ROP (rho of plants) guanine nucleotide exchange factor 14 |
-0.152 |
-0.009 |
| 130 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
-0.151 |
0.184 |
| 131 |
264864_at |
AT1G24310
|
unknown |
-0.150 |
0.017 |
| 132 |
257143_at |
AT3G20110
|
CYP705A20, cytochrome P450, family 705, subfamily A, polypeptide 20 |
-0.150 |
-0.008 |
| 133 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
-0.148 |
0.016 |