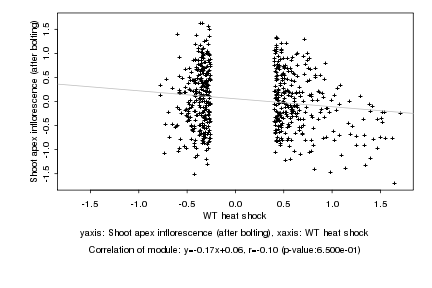
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
WT heat shock |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
253884_at |
AT4G27670
|
HSP21, heat shock protein 21 |
1.704 |
-0.248 |
| 2 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
1.642 |
-1.692 |
| 3 |
258695_at |
AT3G09640
|
APX1B, ASCORBATE PEROXIDASE 1B, APX2, ascorbate peroxidase 2 |
1.622 |
-0.759 |
| 4 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
1.545 |
-0.763 |
| 5 |
250624_at |
AT5G07330
|
unknown |
1.537 |
-0.219 |
| 6 |
255811_at |
AT4G10250
|
ATHSP22.0 |
1.518 |
-0.218 |
| 7 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
1.514 |
-0.430 |
| 8 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
1.508 |
-0.337 |
| 9 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
1.496 |
-0.730 |
| 10 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
1.467 |
-0.369 |
| 11 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
1.460 |
-0.957 |
| 12 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
1.422 |
-0.782 |
| 13 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
1.409 |
-0.088 |
| 14 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
1.392 |
-1.173 |
| 15 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
1.390 |
-0.190 |
| 16 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
1.387 |
-0.057 |
| 17 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
1.344 |
-1.326 |
| 18 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
1.335 |
-0.901 |
| 19 |
259913_at |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
1.331 |
-0.687 |
| 20 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
1.324 |
-0.364 |
| 21 |
244931_at |
ATMG00630
|
ORF110B |
1.285 |
0.117 |
| 22 |
258827_at |
AT3G07150
|
unknown |
1.248 |
-0.721 |
| 23 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
1.244 |
-0.905 |
| 24 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
1.211 |
-0.514 |
| 25 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
1.188 |
-0.669 |
| 26 |
257323_at |
ATMG01200
|
ORF294 |
1.180 |
0.011 |
| 27 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
1.160 |
-0.456 |
| 28 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
1.137 |
-1.381 |
| 29 |
253689_at |
AT4G29770
|
unknown |
1.096 |
-1.103 |
| 30 |
251130_at |
AT5G01180
|
AtNPF8.2, ATPTR5, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 5, NPF8.2, NRT1/ PTR family 8.2, PTR5, peptide transporter 5 |
1.078 |
0.353 |
| 31 |
245243_at |
AT1G44414
|
unknown |
1.072 |
-0.698 |
| 32 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
1.070 |
-0.048 |
| 33 |
254263_at |
AT4G23493
|
unknown |
1.054 |
0.290 |
| 34 |
263164_at |
AT1G03070
|
AtLFG4, LFG4, LIFEGUARD 4 |
1.038 |
-0.184 |
| 35 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
1.018 |
0.141 |
| 36 |
250036_at |
AT5G18340
|
[ARM repeat superfamily protein] |
1.015 |
-0.807 |
| 37 |
256518_at |
AT1G66080
|
unknown |
1.011 |
-0.613 |
| 38 |
247447_at |
AT5G62730
|
[Major facilitator superfamily protein] |
0.989 |
0.033 |
| 39 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
0.974 |
-1.457 |
| 40 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
0.969 |
-0.211 |
| 41 |
254878_at |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
0.950 |
-0.127 |
| 42 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.940 |
-0.733 |
| 43 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.937 |
-0.130 |
| 44 |
264402_at |
AT2G25140
|
CLPB-M, CASEIN LYTIC PROTEINASE B-M, CLPB4, casein lytic proteinase B4, HSP98.7, HEAT SHOCK PROTEIN 98.7 |
0.930 |
0.799 |
| 45 |
250162_at |
AT5G15250
|
ATFTSH6, FTSH6, FTSH protease 6 |
0.920 |
-0.322 |
| 46 |
255485_at |
AT4G02550
|
unknown |
0.915 |
0.475 |
| 47 |
251166_at |
AT3G63350
|
AT-HSFA7B, HSFA7B, HEAT SHOCK TRANSCRIPTION FACTOR A7B |
0.910 |
-0.764 |
| 48 |
259268_at |
AT3G01070
|
AtENODL16, ENODL16, early nodulin-like protein 16 |
0.902 |
0.094 |
| 49 |
265242_at |
AT2G07705
|
unknown |
0.892 |
0.170 |
| 50 |
256363_at |
AT1G66510
|
[AAR2 protein family] |
0.882 |
-0.163 |
| 51 |
246395_at |
AT1G58170
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.880 |
0.547 |
| 52 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
0.871 |
-0.208 |
| 53 |
250910_at |
AT5G03720
|
AT-HSFA3, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A3, HSFA3, heat shock transcription factor A3 |
0.860 |
0.207 |
| 54 |
257337_at |
ATMG00060
|
NAD5, NADH DEHYDROGENASE SUBUNIT 5, NAD5.3, NADH DEHYDROGENASE SUBUNIT 5.3, NAD5C, NADH dehydrogenase subunit 5C |
0.858 |
-0.366 |
| 55 |
260917_at |
AT1G02700
|
unknown |
0.852 |
0.211 |
| 56 |
262014_at |
AT1G35660
|
unknown |
0.848 |
0.041 |
| 57 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.828 |
0.502 |
| 58 |
258794_at |
AT3G04710
|
TPR10, tetratricopeptide repeat 10 |
0.821 |
0.687 |
| 59 |
258336_at |
AT3G16050
|
A37, ATPDX1.2, ARABIDOPSIS THALIANA PYRIDOXINE BIOSYNTHESIS 1.2, PDX1.2, pyridoxine biosynthesis 1.2 |
0.820 |
0.550 |
| 60 |
248215_at |
AT5G53680
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.815 |
-1.393 |
| 61 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
0.804 |
-0.558 |
| 62 |
267036_at |
AT2G38465
|
unknown |
0.801 |
-0.896 |
| 63 |
245789_at |
AT1G32090
|
[early-responsive to dehydration stress protein (ERD4)] |
0.797 |
0.025 |
| 64 |
260668_at |
AT1G19530
|
unknown |
0.796 |
-0.327 |
| 65 |
245272_at |
AT4G17250
|
unknown |
0.786 |
-0.796 |
| 66 |
257118_at |
AT3G20180
|
[Copper transport protein family] |
0.786 |
0.165 |
| 67 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.785 |
-0.287 |
| 68 |
244902_at |
ATMG00650
|
NAD4L, NADH dehydrogenase subunit 4L |
0.784 |
0.123 |
| 69 |
248831_at |
AT5G47160
|
[YDG/SRA domain-containing protein] |
0.777 |
-1.018 |
| 70 |
257321_at |
ATMG01130
|
ORF106F |
0.767 |
0.684 |
| 71 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
0.762 |
0.443 |
| 72 |
245903_at |
AT5G11100
|
ATSYTD, NTMC2T2.2, NTMC2TYPE2.2, SYT4, synaptotagmin 4, SYTD |
0.760 |
-1.056 |
| 73 |
265483_at |
AT2G15790
|
CYP40, CYCLOPHILIN 40, SQN, SQUINT |
0.759 |
0.140 |
| 74 |
246733_at |
AT5G27660
|
DEG14, degradation of periplasmic proteins 14 |
0.757 |
0.909 |
| 75 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.755 |
0.689 |
| 76 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.745 |
0.870 |
| 77 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
0.741 |
1.010 |
| 78 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.739 |
0.422 |
| 79 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
0.726 |
-0.108 |
| 80 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.724 |
0.784 |
| 81 |
259037_at |
AT3G09350
|
Fes1A, Fes1A |
0.720 |
-0.228 |
| 82 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.719 |
-0.800 |
| 83 |
254076_at |
AT4G25340
|
ATFKBP53, FKBP53, FK506 BINDING PROTEIN 53 |
0.709 |
1.306 |
| 84 |
257035_at |
AT3G19270
|
CYP707A4, cytochrome P450, family 707, subfamily A, polypeptide 4 |
0.708 |
0.577 |
| 85 |
257822_at |
AT3G25230
|
ATFKBP62, FKBP62, FK506 BINDING PROTEIN 62, ROF1, rotamase FKBP 1 |
0.703 |
0.938 |
| 86 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.701 |
0.006 |
| 87 |
254043_at |
AT4G25990
|
CIL |
0.699 |
-0.369 |
| 88 |
254574_at |
AT4G19430
|
unknown |
0.694 |
-0.487 |
| 89 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.691 |
0.200 |
| 90 |
256131_at |
AT1G13600
|
AtbZIP58, basic leucine-zipper 58, bZIP58, basic leucine-zipper 58 |
0.685 |
-0.656 |
| 91 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
0.684 |
-0.684 |
| 92 |
264661_at |
AT1G09950
|
RAS1, RESPONSE TO ABA AND SALT 1 |
0.677 |
-0.532 |
| 93 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
0.673 |
-1.089 |
| 94 |
261081_at |
AT1G07350
|
SR45a, serine/arginine rich-like protein 45a |
0.673 |
0.114 |
| 95 |
260025_at |
AT1G30070
|
[SGS domain-containing protein] |
0.669 |
0.118 |
| 96 |
261086_at |
AT1G17460
|
TRFL3, TRF-like 3 |
0.664 |
-0.364 |
| 97 |
252946_at |
AT4G39235
|
unknown |
0.659 |
0.200 |
| 98 |
267627_at |
AT2G42270
|
[U5 small nuclear ribonucleoprotein helicase] |
0.658 |
-0.181 |
| 99 |
261340_at |
AT1G35730
|
APUM9, pumilio 9, PUM9, pumilio 9 |
0.657 |
-0.598 |
| 100 |
244950_at |
ATMG00160
|
COX2, cytochrome oxidase 2 |
0.655 |
0.308 |
| 101 |
246018_at |
AT5G10695
|
unknown |
0.655 |
-0.520 |
| 102 |
246612_at |
AT5G35320
|
unknown |
0.649 |
-0.367 |
| 103 |
264019_at |
AT2G21130
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.641 |
0.753 |
| 104 |
264389_at |
AT1G11960
|
[ERD (early-responsive to dehydration stress) family protein] |
0.640 |
-0.104 |
| 105 |
253113_at |
AT4G35985
|
[Senescence/dehydration-associated protein-related] |
0.636 |
-0.082 |
| 106 |
263732_at |
AT1G59980
|
ARL2, ARG1-like 2, ATDJC39, GPS4, gravity persistence signal 4 |
0.633 |
1.000 |
| 107 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
0.633 |
0.524 |
| 108 |
247119_at |
AT5G65900
|
[DEA(D/H)-box RNA helicase family protein] |
0.633 |
-0.236 |
| 109 |
255651_at |
AT4G00940
|
AtDOF4.1, DOF4.1, DNA binding with one finger 4.1, ITD1, INTERCELLULAR TRAFFICKING DOF 1 |
0.631 |
-0.077 |
| 110 |
245293_at |
AT4G16660
|
[heat shock protein 70 (Hsp 70) family protein] |
0.629 |
0.180 |
| 111 |
261655_at |
AT1G01940
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.628 |
0.982 |
| 112 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.626 |
-0.293 |
| 113 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.624 |
0.189 |
| 114 |
267462_at |
AT2G33735
|
[Chaperone DnaJ-domain superfamily protein] |
0.620 |
-0.661 |
| 115 |
267521_at |
AT2G30480
|
unknown |
0.612 |
-0.234 |
| 116 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.611 |
0.006 |
| 117 |
250502_at |
AT5G09590
|
HSC70-5, HEAT SHOCK COGNATE, MTHSC70-2, mitochondrial HSO70 2 |
0.611 |
-0.068 |
| 118 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.609 |
0.067 |
| 119 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
0.608 |
-0.093 |
| 120 |
264968_at |
AT1G67360
|
[Rubber elongation factor protein (REF)] |
0.604 |
0.423 |
| 121 |
260522_x_at |
AT2G41730
|
unknown |
0.602 |
-1.036 |
| 122 |
244903_at |
ATMG00660
|
ORF149 |
0.602 |
0.568 |
| 123 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
0.600 |
-0.201 |
| 124 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
0.599 |
0.587 |
| 125 |
244904_at |
ATMG00670
|
ORF275 |
0.590 |
0.176 |
| 126 |
248495_at |
AT5G50780
|
[Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase family protein] |
0.590 |
-0.197 |
| 127 |
248596_at |
AT5G49330
|
ATMYB111, ARABIDOPSIS MYB DOMAIN PROTEIN 111, MYB111, myb domain protein 111, PFG3, PRODUCTION OF FLAVONOL GLYCOSIDES 3 |
0.588 |
0.929 |
| 128 |
258406_at |
AT3G17611
|
ATRBL10, RHOMBOID-like protein 10, ATRBL14, RHOMBOID-like protein 14, RBL10, RHOMBOID-like protein 10, RBL14, RHOMBOID-like protein 14 |
0.585 |
-0.184 |
| 129 |
259169_at |
AT3G03520
|
NPC3, non-specific phospholipase C3 |
0.583 |
-0.207 |
| 130 |
264890_at |
AT1G23180
|
[ARM repeat superfamily protein] |
0.582 |
-0.164 |
| 131 |
264338_at |
AT1G70300
|
KUP6, K+ uptake permease 6 |
0.582 |
-0.296 |
| 132 |
261167_at |
AT1G04980
|
ATPDI10, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 10, ATPDIL2-2, PDI-like 2-2, PDI10, PROTEIN DISULFIDE ISOMERASE, PDIL2-2, PDI-like 2-2 |
0.578 |
0.348 |
| 133 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.578 |
-0.123 |
| 134 |
264814_at |
AT2G17900
|
ASHR1, ASH1-related 1, SDG37, SET domain group 37 |
0.576 |
0.446 |
| 135 |
262691_at |
AT1G62740
|
Hop2, Hop2 |
0.576 |
0.862 |
| 136 |
257332_at |
ATMG01350
|
ORF145C |
0.576 |
-0.432 |
| 137 |
245228_at |
AT3G29810
|
COBL2, COBRA-like protein 2 precursor |
0.575 |
0.630 |
| 138 |
244906_at |
ATMG00690
|
ORF240A |
0.571 |
-0.230 |
| 139 |
246386_at |
AT1G77410
|
BGAL16, beta-galactosidase 16 |
0.570 |
-0.160 |
| 140 |
257317_at |
ATMG01060
|
ORF107G |
0.570 |
-0.809 |
| 141 |
244921_s_at |
ATMG01000
|
ORF114 |
0.570 |
0.116 |
| 142 |
264142_at |
AT1G78930
|
[Mitochondrial transcription termination factor family protein] |
0.567 |
-0.076 |
| 143 |
267336_at |
AT2G19310
|
[HSP20-like chaperones superfamily protein] |
0.567 |
-1.195 |
| 144 |
255077_at |
AT4G09150
|
[T-complex protein 11] |
0.565 |
0.688 |
| 145 |
246125_at |
AT5G19875
|
unknown |
0.560 |
-0.335 |
| 146 |
248258_at |
AT5G53400
|
BOB1, BOBBER1 |
0.558 |
0.189 |
| 147 |
266408_at |
AT2G38520
|
[copia-like retrotransposon family, has a 8.4e-60 P-value blast match to dbj|BAA78427.1| polyprotein (AtRE2-2) (Arabidopsis thaliana) (Ty1_Copia-element)] |
0.552 |
-0.506 |
| 148 |
256430_at |
AT3G11020
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2B, DRE/CRT-binding protein 2B |
0.550 |
-0.921 |
| 149 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.549 |
-0.759 |
| 150 |
253859_at |
AT4G27657
|
unknown |
0.548 |
-0.116 |
| 151 |
262322_at |
AT1G27590
|
unknown |
0.545 |
-0.463 |
| 152 |
251049_at |
AT5G02430
|
[Transducin/WD40 repeat-like superfamily protein] |
0.541 |
0.210 |
| 153 |
258979_at |
AT3G09440
|
[Heat shock protein 70 (Hsp 70) family protein] |
0.541 |
0.634 |
| 154 |
253687_at |
AT4G29520
|
[LOCATED IN: endoplasmic reticulum, plasma membrane] |
0.541 |
0.136 |
| 155 |
261300_at |
AT1G48560
|
unknown |
0.538 |
-0.294 |
| 156 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.537 |
0.906 |
| 157 |
262814_at |
AT1G11660
|
[heat shock protein 70 (Hsp 70) family protein] |
0.537 |
0.232 |
| 158 |
248463_at |
AT5G51130
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.537 |
0.700 |
| 159 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.536 |
-0.063 |
| 160 |
254211_at |
AT4G23570
|
SGT1A |
0.535 |
0.467 |
| 161 |
265147_at |
AT1G51380
|
[DEA(D/H)-box RNA helicase family protein] |
0.531 |
-0.792 |
| 162 |
264331_at |
AT1G04130
|
AtTPR2, TPR2, tetratricopeptide repeat 2 |
0.531 |
-0.528 |
| 163 |
264522_at |
AT1G10050
|
[glycosyl hydrolase family 10 protein / carbohydrate-binding domain-containing protein] |
0.523 |
-0.268 |
| 164 |
261844_at |
AT1G15940
|
[Tudor/PWWP/MBT superfamily protein] |
0.523 |
-0.045 |
| 165 |
259248_at |
AT3G07770
|
AtHsp90-6, HEAT SHOCK PROTEIN 90-6, AtHsp90.6, HEAT SHOCK PROTEIN 90.6, Hsp89.1, HEAT SHOCK PROTEIN 89.1 |
0.522 |
1.208 |
| 166 |
253064_at |
AT4G37730
|
AtbZIP7, basic leucine-zipper 7, bZIP7, basic leucine-zipper 7 |
0.521 |
-0.223 |
| 167 |
261219_at |
AT1G19980
|
[cytomatrix protein-related] |
0.518 |
0.757 |
| 168 |
257336_at |
ATMG01410
|
ORF204, open reading frame 204 |
0.518 |
-0.659 |
| 169 |
261610_at |
AT1G49560
|
[Homeodomain-like superfamily protein] |
0.517 |
-0.214 |
| 170 |
263515_at |
AT2G21640
|
unknown |
0.511 |
0.213 |
| 171 |
265151_at |
AT1G51340
|
[MATE efflux family protein] |
0.509 |
-1.205 |
| 172 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.508 |
-0.106 |
| 173 |
251563_at |
AT3G57880
|
[Calcium-dependent lipid-binding (CaLB domain) plant phosphoribosyltransferase family protein] |
0.506 |
0.071 |
| 174 |
246770_at |
AT5G27460
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.505 |
0.713 |
| 175 |
253987_at |
AT4G26270
|
PFK3, phosphofructokinase 3 |
0.505 |
0.696 |
| 176 |
256663_at |
AT3G12050
|
[Aha1 domain-containing protein] |
0.505 |
0.382 |
| 177 |
248045_at |
AT5G56030
|
AtHsp90.2, HEAT SHOCK PROTEIN 90.2, ERD8, EARLY-RESPONSIVE TO DEHYDRATION 8, HSP81-2, heat shock protein 81-2, HSP81.2, heat shock protein 81.2, HSP90.2, HEAT SHOCK PROTEIN 90.2 |
0.502 |
0.486 |
| 178 |
244901_at |
ATMG00640
|
ORF25 |
0.501 |
-0.040 |
| 179 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
0.501 |
0.542 |
| 180 |
261839_at |
AT1G16040
|
unknown |
0.500 |
0.169 |
| 181 |
266806_at |
AT2G30000
|
[PHF5-like protein] |
0.499 |
0.199 |
| 182 |
267034_at |
AT2G38310
|
PYL4, PYR1-like 4, RCAR10, regulatory components of ABA receptor 10 |
0.497 |
0.582 |
| 183 |
266046_at |
AT2G07728
|
unknown |
0.496 |
-0.335 |
| 184 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
0.495 |
-0.217 |
| 185 |
247187_at |
AT5G65490
|
unknown |
0.494 |
0.895 |
| 186 |
245313_at |
AT4G15420
|
[Ubiquitin fusion degradation UFD1 family protein] |
0.492 |
0.019 |
| 187 |
267073_at |
AT2G41160
|
[Ubiquitin-associated (UBA) protein] |
0.490 |
0.191 |
| 188 |
256905_at |
AT3G23990
|
HSP60, heat shock protein 60, HSP60-3B, HEAT SHOCK PROTEIN 60-3B |
0.489 |
0.736 |
| 189 |
265442_at |
AT2G20940
|
unknown |
0.489 |
0.603 |
| 190 |
248774_at |
AT5G47830
|
unknown |
0.489 |
0.288 |
| 191 |
258830_at |
AT3G07090
|
[PPPDE putative thiol peptidase family protein] |
0.486 |
0.439 |
| 192 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.483 |
-0.436 |
| 193 |
264358_at |
AT1G03180
|
unknown |
0.482 |
0.487 |
| 194 |
261485_at |
AT1G14360
|
ATUTR3, UTR3, UDP-galactose transporter 3 |
0.481 |
0.573 |
| 195 |
248228_at |
AT5G53800
|
unknown |
0.479 |
-0.003 |
| 196 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
0.477 |
-0.803 |
| 197 |
263128_at |
AT1G78600
|
BBX22, B-box domain protein 22, DBB3, DOUBLE B-BOX 3, LZF1, light-regulated zinc finger protein 1, STH3, SALT TOLERANCE HOMOLOG 3 |
0.476 |
1.093 |
| 198 |
249256_at |
AT5G41620
|
unknown |
0.475 |
-0.538 |
| 199 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
0.474 |
-0.130 |
| 200 |
247509_at |
AT5G62020
|
AT-HSFB2A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR B2A, HSFB2A, heat shock transcription factor B2A |
0.474 |
-0.325 |
| 201 |
264357_at |
AT1G03360
|
ATRRP4, ribosomal RNA processing 4, RRP4, ribosomal RNA processing 4 |
0.474 |
0.134 |
| 202 |
249233_at |
AT5G42150
|
[Glutathione S-transferase family protein] |
0.473 |
0.129 |
| 203 |
262706_at |
AT1G16280
|
AtRH36, Arabidopsis thaliana RNA helicase 36, RH36, RNA helicase 36, SWA3, SLOW WALKER 3 |
0.472 |
1.221 |
| 204 |
260425_at |
AT1G72440
|
EDA25, embryo sac development arrest 25, SWA2, SLOW WALKER2 |
0.471 |
1.073 |
| 205 |
260987_at |
AT1G53590
|
NTMC2T6.1, NTMC2TYPE6.1 |
0.471 |
-0.491 |
| 206 |
258871_at |
AT3G03060
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.471 |
-0.609 |
| 207 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.469 |
0.559 |
| 208 |
253249_at |
AT4G34680
|
GATA3, GATA transcription factor 3 |
0.468 |
-0.758 |
| 209 |
255509_at |
AT4G02210
|
unknown |
0.467 |
1.022 |
| 210 |
257719_at |
AT3G18440
|
ALMT9, aluminum-activated malate transporter 9, AtALMT9, aluminum-activated malate transporter 9 |
0.465 |
0.395 |
| 211 |
255787_at |
AT2G33590
|
AtCRL1, CRL1, CCR(Cinnamoyl coA:NADP oxidoreductase)-like 1 |
0.465 |
-0.241 |
| 212 |
247104_at |
AT5G66620
|
DAR6, DA1-related protein 6 |
0.463 |
-0.275 |
| 213 |
266836_at |
AT2G26000
|
BRIZ2, BRAP2 RING ZnF UBP domain-containing protein 2 |
0.462 |
-0.233 |
| 214 |
263897_at |
AT2G21940
|
ATSK1, SK1, shikimate kinase 1 |
0.460 |
0.880 |
| 215 |
245828_at |
AT1G57820
|
ORTH2, ORTHRUS 2, VIM1, VARIANT IN METHYLATION 1 |
0.460 |
0.336 |
| 216 |
250087_at |
AT5G17270
|
[Protein prenylyltransferase superfamily protein] |
0.459 |
-0.487 |
| 217 |
254663_at |
AT4G18290
|
KAT2, potassium channel in Arabidopsis thaliana 2 |
0.457 |
-0.872 |
| 218 |
258805_at |
AT3G04010
|
[O-Glycosyl hydrolases family 17 protein] |
0.457 |
-0.902 |
| 219 |
254402_at |
AT4G21310
|
unknown |
0.455 |
-0.240 |
| 220 |
246554_at |
AT5G15450
|
APG6, ALBINO AND PALE GREEN 6, AtCLPB3, CLPB-P, CASEIN LYTIC PROTEINASE B-P, CLPB3, casein lytic proteinase B3 |
0.454 |
0.749 |
| 221 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
0.454 |
-0.385 |
| 222 |
248136_at |
AT5G54910
|
[DEA(D/H)-box RNA helicase family protein] |
0.452 |
-0.624 |
| 223 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
0.451 |
-0.412 |
| 224 |
258262_at |
AT3G15770
|
unknown |
0.450 |
-0.254 |
| 225 |
247350_at |
AT5G63830
|
[HIT-type Zinc finger family protein] |
0.450 |
-0.035 |
| 226 |
255382_at |
AT4G03430
|
EMB2770, EMBRYO DEFECTIVE 2770, STA1, STABILIZED 1 |
0.450 |
0.680 |
| 227 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
0.449 |
-0.376 |
| 228 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
0.446 |
-0.806 |
| 229 |
267483_at |
AT2G02810
|
ATUTR1, UDP-GALACTOSE TRANSPORTER 1, UTR1, UDP-galactose transporter 1 |
0.445 |
0.845 |
| 230 |
244958_at |
ATMG00450
|
ORF106B |
0.444 |
-0.275 |
| 231 |
251522_at |
AT3G59430
|
unknown |
0.444 |
-0.279 |
| 232 |
249365_at |
AT5G40600
|
unknown |
0.443 |
0.236 |
| 233 |
254190_at |
AT4G23885
|
unknown |
0.443 |
0.084 |
| 234 |
247567_at |
AT5G61190
|
[putative endonuclease or glycosyl hydrolase with C2H2-type zinc finger domain] |
0.442 |
-0.574 |
| 235 |
258522_at |
AT3G06660
|
[PAPA-1-like family protein / zinc finger (HIT type) family protein] |
0.441 |
0.035 |
| 236 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.441 |
0.723 |
| 237 |
262656_at |
AT1G14200
|
[RING/U-box superfamily protein] |
0.441 |
-0.023 |
| 238 |
264704_at |
AT1G70090
|
GATL9, GALACTURONOSYLTRANSFERASE-LIKE 9, LGT8, glucosyl transferase family 8 |
0.440 |
0.190 |
| 239 |
257814_at |
AT3G25110
|
AtFaTA, fatA acyl-ACP thioesterase, FaTA, fatA acyl-ACP thioesterase |
0.439 |
0.937 |
| 240 |
265935_at |
AT2G19580
|
TET2, tetraspanin2 |
0.438 |
-0.499 |
| 241 |
257931_at |
AT3G17030
|
[Nucleic acid-binding proteins superfamily] |
0.437 |
0.260 |
| 242 |
265061_at |
AT1G61640
|
[Protein kinase superfamily protein] |
0.436 |
0.023 |
| 243 |
256983_at |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
0.436 |
0.744 |
| 244 |
247792_at |
AT5G58787
|
[RING/U-box superfamily protein] |
0.434 |
0.034 |
| 245 |
265991_at |
AT2G24120
|
PDE319, PIGMENT DEFECTIVE 319, SCA3, SCABRA 3 |
0.434 |
0.475 |
| 246 |
253166_at |
AT4G35290
|
ATGLR3.2, ATGLUR2, GLR3.2, GLUTAMATE RECEPTOR 3.2, GLUR2, glutamate receptor 2 |
0.432 |
-0.823 |
| 247 |
262098_at |
AT1G56170
|
ATHAP5B, HAP5B, NF-YC2, nuclear factor Y, subunit C2 |
0.432 |
0.559 |
| 248 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
0.431 |
-0.740 |
| 249 |
265930_at |
AT2G18510
|
emb2444, embryo defective 2444 |
0.431 |
1.322 |
| 250 |
260252_at |
AT1G74240
|
[Mitochondrial substrate carrier family protein] |
0.430 |
0.161 |
| 251 |
253712_at |
AT4G29330
|
DER1, DERLIN-1 |
0.428 |
0.786 |
| 252 |
250332_at |
AT5G11680
|
unknown |
0.428 |
-0.348 |
| 253 |
252586_at |
AT3G45610
|
DOF6, DOF transcription factor 6 |
0.428 |
-0.353 |
| 254 |
264726_at |
AT1G22985
|
CRF7, cytokinin response factor 7 |
0.428 |
-0.307 |
| 255 |
259540_at |
AT1G20640
|
NLP4, NIN-like protein 4 |
0.426 |
-0.389 |
| 256 |
253456_at |
AT4G32050
|
[neurochondrin family protein] |
0.425 |
-0.217 |
| 257 |
263921_at |
AT2G36460
|
FBA6, fructose-bisphosphate aldolase 6 |
0.424 |
1.335 |
| 258 |
250103_at |
AT5G16600
|
AtMYB43, myb domain protein 43, MYB43, myb domain protein 43 |
0.423 |
0.079 |
| 259 |
250841_at |
AT5G04610
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.422 |
-0.792 |
| 260 |
258812_at |
AT3G03950
|
ECT1, evolutionarily conserved C-terminal region 1 |
0.422 |
1.047 |
| 261 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
0.422 |
0.540 |
| 262 |
260283_at |
AT1G80480
|
PTAC17, plastid transcriptionally active 17 |
0.422 |
1.101 |
| 263 |
245256_at |
AT4G15090
|
FAR1, FAR-RED IMPAIRED RESPONSE 1 |
0.421 |
-0.065 |
| 264 |
250899_at |
AT5G03340
|
AtCDC48C, cell division cycle 48C |
0.421 |
1.310 |
| 265 |
250127_at |
AT5G16380
|
unknown |
0.421 |
-0.402 |
| 266 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.420 |
0.146 |
| 267 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.419 |
-0.821 |
| 268 |
258460_at |
AT3G17330
|
ECT6, evolutionarily conserved C-terminal region 6 |
0.418 |
0.617 |
| 269 |
265326_at |
AT2G18220
|
[Noc2p family] |
0.418 |
0.703 |
| 270 |
252305_at |
AT3G49240
|
emb1796, embryo defective 1796 |
0.417 |
0.499 |
| 271 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
0.416 |
0.129 |
| 272 |
262558_at |
AT1G31335
|
unknown |
0.416 |
1.171 |
| 273 |
257333_at |
ATMG01360
|
COX1, cytochrome oxidase |
0.416 |
0.097 |
| 274 |
258177_at |
AT3G21660
|
[UBX domain-containing protein] |
0.416 |
-0.591 |
| 275 |
246309_at |
AT3G51790
|
AtCCME, A. thaliana homolog of E. coli CcmE, ATG1, transmembrane protein G1P-related 1, G1, transmembrane protein G1P-related 1 |
0.416 |
0.455 |
| 276 |
257487_at |
AT1G71850
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
0.416 |
-0.191 |
| 277 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
0.415 |
-0.331 |
| 278 |
251078_at |
AT5G01990
|
[Auxin efflux carrier family protein] |
0.415 |
0.088 |
| 279 |
262677_at |
AT1G75860
|
unknown |
0.415 |
1.132 |
| 280 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
0.414 |
0.070 |
| 281 |
267305_at |
AT2G30070
|
ATKT1, potassium transporter 1, ATKT1P, ATKUP1, KT1, potassium transporter 1, KUP1, POTASSIUM UPTAKE TRANSPORTER 1 |
0.413 |
0.635 |
| 282 |
259325_at |
AT3G05320
|
[O-fucosyltransferase family protein] |
0.413 |
-0.584 |
| 283 |
259958_at |
AT1G53730
|
SRF6, STRUBBELIG-receptor family 6 |
0.412 |
1.011 |
| 284 |
264787_at |
AT2G17840
|
ERD7, EARLY-RESPONSIVE TO DEHYDRATION 7 |
0.410 |
0.223 |
| 285 |
264583_at |
AT1G05170
|
[Galactosyltransferase family protein] |
0.408 |
-0.327 |
| 286 |
252485_at |
AT3G46530
|
RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13 |
0.408 |
-0.662 |
| 287 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.407 |
-1.054 |
| 288 |
251469_at |
AT3G59530
|
LAP3, LESS ADHERENT POLLEN 3 |
0.407 |
-0.625 |
| 289 |
258939_at |
AT3G10020
|
unknown |
0.407 |
0.327 |
| 290 |
250930_at |
AT5G03160
|
ATP58IPK, homolog of mamallian P58IPK, P58IPK, homolog of mamallian P58IPK |
0.406 |
0.226 |
| 291 |
247657_at |
AT5G59845
|
[Gibberellin-regulated family protein] |
0.406 |
-0.419 |
| 292 |
266574_at |
AT2G23890
|
[HAD-superfamily hydrolase, subfamily IG, 5'-nucleotidase] |
0.405 |
-0.495 |
| 293 |
247287_at |
AT5G64230
|
unknown |
0.403 |
0.228 |
| 294 |
257780_at |
AT3G27100
|
unknown |
0.403 |
-0.147 |
| 295 |
267190_at |
AT2G44170
|
ATNMT2, ARABIDOPSIS N-MYRISTOYLTRANSFERASE 2, NMT2, N-myristoyltransferase 2 |
0.401 |
-0.058 |
| 296 |
254526_at |
AT4G19600
|
CYCT1;4 |
0.400 |
0.850 |
| 297 |
249577_at |
AT5G37710
|
[alpha/beta-Hydrolases superfamily protein] |
0.400 |
0.757 |
| 298 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
0.399 |
-0.406 |
| 299 |
254778_at |
AT4G12750
|
[Homeodomain-like transcriptional regulator] |
0.399 |
0.896 |
| 300 |
256734_at |
AT3G29390
|
RIK, RS2-interacting KH protein |
0.398 |
1.071 |
| 301 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
-0.783 |
0.138 |
| 302 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
-0.779 |
0.348 |
| 303 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
-0.742 |
-1.076 |
| 304 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.729 |
-0.473 |
| 305 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
-0.720 |
0.477 |
| 306 |
248889_at |
AT5G46230
|
unknown |
-0.699 |
-0.222 |
| 307 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
-0.684 |
-0.746 |
| 308 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.654 |
-0.465 |
| 309 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.652 |
0.285 |
| 310 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.622 |
-0.529 |
| 311 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
-0.617 |
-0.518 |
| 312 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.608 |
-0.490 |
| 313 |
258480_at |
AT3G02640
|
unknown |
-0.608 |
1.399 |
| 314 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.607 |
-0.106 |
| 315 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.595 |
-1.032 |
| 316 |
247444_at |
AT5G62630
|
HIPL2, hipl2 protein precursor |
-0.589 |
0.240 |
| 317 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.586 |
-0.162 |
| 318 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
-0.586 |
-0.959 |
| 319 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
-0.584 |
0.179 |
| 320 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.583 |
0.535 |
| 321 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.581 |
0.928 |
| 322 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
-0.579 |
0.195 |
| 323 |
257074_at |
AT3G19660
|
unknown |
-0.572 |
-0.782 |
| 324 |
265083_at |
AT1G03820
|
unknown |
-0.571 |
0.175 |
| 325 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
-0.559 |
-0.230 |
| 326 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
-0.558 |
0.484 |
| 327 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
-0.537 |
-0.922 |
| 328 |
261032_at |
AT1G17430
|
[alpha/beta-Hydrolases superfamily protein] |
-0.532 |
0.204 |
| 329 |
264377_at |
AT2G25060
|
AtENODL14, ENODL14, early nodulin-like protein 14 |
-0.526 |
0.674 |
| 330 |
255596_at |
AT4G01720
|
AtWRKY47, WRKY47 |
-0.523 |
0.189 |
| 331 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
-0.522 |
0.311 |
| 332 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.520 |
-0.168 |
| 333 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.514 |
-0.165 |
| 334 |
253165_at |
AT4G35320
|
unknown |
-0.512 |
-0.281 |
| 335 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
-0.509 |
-0.976 |
| 336 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.504 |
-0.415 |
| 337 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
-0.503 |
-0.311 |
| 338 |
253779_at |
AT4G28490
|
HAE, HAESA, RLK5, RECEPTOR-LIKE PROTEIN KINASE 5 |
-0.495 |
-0.325 |
| 339 |
256021_at |
AT1G58270
|
ZW9 |
-0.494 |
-0.195 |
| 340 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.494 |
-0.003 |
| 341 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
-0.490 |
0.380 |
| 342 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.490 |
-0.237 |
| 343 |
262646_at |
AT1G62800
|
ASP4, aspartate aminotransferase 4 |
-0.487 |
-0.377 |
| 344 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.483 |
0.266 |
| 345 |
245896_at |
AT5G09250
|
KIWI |
-0.483 |
0.508 |
| 346 |
247651_at |
AT5G59870
|
HTA6, histone H2A 6 |
-0.482 |
0.696 |
| 347 |
260754_at |
AT1G49000
|
unknown |
-0.480 |
-0.139 |
| 348 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
-0.479 |
-0.055 |
| 349 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
-0.478 |
-0.375 |
| 350 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
-0.475 |
0.456 |
| 351 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
-0.467 |
0.033 |
| 352 |
247218_at |
AT5G65010
|
ASN2, asparagine synthetase 2 |
-0.467 |
-0.329 |
| 353 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.466 |
0.239 |
| 354 |
267590_at |
AT2G39700
|
ATEXP4, ATEXPA4, expansin A4, ATHEXP ALPHA 1.6, EXPA4, expansin A4 |
-0.461 |
0.604 |
| 355 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
-0.460 |
-0.494 |
| 356 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.459 |
-0.507 |
| 357 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
-0.459 |
-0.780 |
| 358 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.456 |
-0.106 |
| 359 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.454 |
-0.599 |
| 360 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
-0.452 |
0.439 |
| 361 |
250690_at |
AT5G06530
|
ABCG22, ATP-binding cassette G22, AtABCG22, Arabidopsis thaliana ATP-binding cassette G22 |
-0.446 |
-0.074 |
| 362 |
245445_at |
AT4G16750
|
[Integrase-type DNA-binding superfamily protein] |
-0.445 |
-0.570 |
| 363 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
-0.442 |
-0.612 |
| 364 |
264262_at |
AT1G09200
|
H3.1, histone 3.1 |
-0.440 |
0.855 |
| 365 |
259014_at |
AT3G07320
|
[O-Glycosyl hydrolases family 17 protein] |
-0.437 |
0.218 |
| 366 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.435 |
0.451 |
| 367 |
262935_at |
AT1G79410
|
AtOCT5, organic cation/carnitine transporter5, OCT5, organic cation/carnitine transporter5 |
-0.432 |
-0.499 |
| 368 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.432 |
-0.548 |
| 369 |
252442_at |
AT3G46940
|
DUT1, DUTP-PYROPHOSPHATASE-LIKE 1 |
-0.431 |
0.457 |
| 370 |
252533_at |
AT3G46110
|
[LOCATED IN: plasma membrane] |
-0.431 |
-0.868 |
| 371 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.431 |
-0.002 |
| 372 |
267550_at |
AT2G32800
|
AP4.3A, LecRK-S.2, L-type lectin receptor kinase S.2 |
-0.429 |
-0.918 |
| 373 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-0.428 |
-0.197 |
| 374 |
252539_at |
AT3G45730
|
unknown |
-0.424 |
-1.496 |
| 375 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
-0.424 |
0.139 |
| 376 |
248968_at |
AT5G45280
|
[Pectinacetylesterase family protein] |
-0.423 |
0.388 |
| 377 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.422 |
-0.203 |
| 378 |
248705_at |
AT5G48520
|
AtAUG3, AUG3, augmin 3 |
-0.420 |
0.445 |
| 379 |
256091_at |
AT1G20693
|
HMG BETA 1, HIGH MOBILITY GROUP BETA 1, HMGB2, high mobility group B2, NFD02, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP D 02, NFD2, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP D 2 |
-0.420 |
0.870 |
| 380 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
-0.420 |
-0.379 |
| 381 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
-0.420 |
-0.794 |
| 382 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.419 |
-1.148 |
| 383 |
264025_at |
AT2G21050
|
LAX2, like AUXIN RESISTANT 2 |
-0.418 |
0.377 |
| 384 |
257147_at |
AT3G27270
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
-0.417 |
0.397 |
| 385 |
263480_at |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-0.417 |
0.111 |
| 386 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.415 |
0.880 |
| 387 |
258222_at |
AT3G15680
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-0.415 |
0.212 |
| 388 |
250560_at |
AT5G08020
|
ATRPA70B, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT B, RPA70B, RPA70-kDa subunit B |
-0.415 |
1.192 |
| 389 |
249167_at |
AT5G42860
|
unknown |
-0.413 |
0.250 |
| 390 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.409 |
0.033 |
| 391 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.408 |
0.719 |
| 392 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.407 |
-0.287 |
| 393 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-0.406 |
0.220 |
| 394 |
245112_at |
AT2G41540
|
GPDHC1 |
-0.406 |
0.670 |
| 395 |
266695_at |
AT2G19810
|
AtOZF1, AtTZF2, OZF1, Oxidation-related Zinc Finger 1, TZF2, tandem zinc finger 2 |
-0.404 |
1.373 |
| 396 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
-0.404 |
-0.962 |
| 397 |
260033_at |
AT1G68760
|
ATNUDT1, ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 1, ATNUDX1, nudix hydrolase 1, NUDX1, nudix hydrolase 1, NUDX1, NUDIX HYDROLASE 1 |
-0.403 |
0.169 |
| 398 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
-0.399 |
-1.111 |
| 399 |
260392_at |
AT1G74030
|
ENO1, enolase 1 |
-0.398 |
0.493 |
| 400 |
260506_at |
AT1G47210
|
CYCA3;2, cyclin-dependent protein kinase 3;2 |
-0.396 |
0.660 |
| 401 |
246518_at |
AT5G15770
|
AtGNA1, glucose-6-phosphate acetyltransferase 1, GNA1, glucose-6-phosphate acetyltransferase 1 |
-0.396 |
-0.284 |
| 402 |
267503_at |
AT2G45600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.395 |
0.481 |
| 403 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
-0.392 |
0.857 |
| 404 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.387 |
0.546 |
| 405 |
248923_at |
AT5G45940
|
AtNUDT11, Arabidopsis thaliana nudix hydrolase homolog 11, AtNUDX11, Arabidopsis thaliana nudix hydrolase homolog 11, NUDT11, nudix hydrolase homolog 11, NUDX11, nudix hydrolase homolog 11 |
-0.387 |
0.686 |
| 406 |
250732_at |
AT5G06480
|
[Immunoglobulin E-set superfamily protein] |
-0.386 |
0.779 |
| 407 |
256785_at |
AT3G13720
|
PRA1.F3, PRENYLATED RAB ACCEPTOR 1.F3, PRA8 |
-0.386 |
1.058 |
| 408 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
-0.385 |
0.392 |
| 409 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.383 |
1.107 |
| 410 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.382 |
-0.239 |
| 411 |
248192_at |
AT5G54140
|
ILL3, IAA-leucine-resistant (ILR1)-like 3 |
-0.381 |
-0.702 |
| 412 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.379 |
0.188 |
| 413 |
262624_at |
AT1G06450
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
-0.379 |
0.081 |
| 414 |
258379_at |
AT3G16700
|
[Fumarylacetoacetate (FAA) hydrolase family] |
-0.378 |
-0.484 |
| 415 |
247169_at |
AT5G65520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.377 |
-0.062 |
| 416 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
-0.375 |
-0.620 |
| 417 |
250586_at |
AT5G07630
|
[lipid transporters] |
-0.373 |
-0.003 |
| 418 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.371 |
0.054 |
| 419 |
261794_at |
AT1G16060
|
ADAP, ARIA-interacting double AP2 domain protein, WRI3, WRINKLED 3 |
-0.369 |
-0.105 |
| 420 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
-0.369 |
-0.602 |
| 421 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
-0.369 |
-0.671 |
| 422 |
264133_at |
AT1G79080
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.368 |
0.634 |
| 423 |
250417_at |
AT5G11230
|
[Nucleotide-sugar transporter family protein] |
-0.368 |
0.071 |
| 424 |
250043_at |
AT5G18430
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.366 |
-0.497 |
| 425 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
-0.366 |
-0.496 |
| 426 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
-0.364 |
1.637 |
| 427 |
252179_at |
AT3G50760
|
GATL2, galacturonosyltransferase-like 2 |
-0.363 |
0.568 |
| 428 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.363 |
0.478 |
| 429 |
247606_at |
AT5G61000
|
ATRPA70D, RPA70D |
-0.362 |
1.165 |
| 430 |
252077_at |
AT3G51720
|
unknown |
-0.362 |
0.756 |
| 431 |
259210_at |
AT3G09150
|
ATHY2, ARABIDOPSIS ELONGATED HYPOCOTYL 2, GUN3, GENOMES UNCOUPLED 3, HY2, ELONGATED HYPOCOTYL 2 |
-0.362 |
0.138 |
| 432 |
251010_at |
AT5G02550
|
unknown |
-0.361 |
0.801 |
| 433 |
247192_at |
AT5G65360
|
H3.1, histone 3.1 |
-0.360 |
1.019 |
| 434 |
260645_at |
AT1G53300
|
TTL1, tetratricopeptide-repeat thioredoxin-like 1 |
-0.360 |
0.865 |
| 435 |
257610_at |
AT3G13810
|
AtIDD11, indeterminate(ID)-domain 11, IDD11, indeterminate(ID)-domain 11 |
-0.359 |
0.677 |
| 436 |
260618_at |
AT1G53230
|
TCP3, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 3 |
-0.359 |
0.301 |
| 437 |
252133_at |
AT3G50900
|
unknown |
-0.358 |
0.256 |
| 438 |
261368_at |
AT1G53070
|
[Legume lectin family protein] |
-0.357 |
0.450 |
| 439 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.357 |
0.417 |
| 440 |
249469_at |
AT5G39320
|
UDG4, UDP-glucose dehydrogenase 4 |
-0.356 |
0.928 |
| 441 |
267618_at |
AT2G26760
|
CYCB1;4, Cyclin B1;4 |
-0.356 |
0.928 |
| 442 |
247800_at |
AT5G58570
|
unknown |
-0.355 |
0.147 |
| 443 |
250696_at |
AT5G06790
|
unknown |
-0.355 |
-0.661 |
| 444 |
254924_at |
AT4G11330
|
ATMPK5, MAP kinase 5, MPK5, MAP kinase 5 |
-0.355 |
-0.645 |
| 445 |
252934_at |
AT4G39120
|
HISN7, HISTIDINE BIOSYNTHESIS 7, IMPL2, myo-inositol monophosphatase like 2 |
-0.355 |
0.136 |
| 446 |
255511_at |
AT4G02075
|
PIT1, pitchoun 1 |
-0.354 |
0.497 |
| 447 |
253533_at |
AT4G31590
|
ATCSLC05, CELLULOSE-SYNTHASE LIKE C5, ATCSLC5, Cellulose-synthase-like C5, CSLC05, CELLULOSE-SYNTHASE LIKE C5, CSLC5, Cellulose-synthase-like C5 |
-0.352 |
-0.283 |
| 448 |
267344_at |
AT2G44230
|
unknown |
-0.351 |
-0.478 |
| 449 |
251986_at |
AT3G53310
|
[AP2/B3-like transcriptional factor family protein] |
-0.351 |
1.082 |
| 450 |
262788_at |
AT1G10690
|
unknown |
-0.350 |
-0.208 |
| 451 |
254119_at |
AT4G24780
|
[Pectin lyase-like superfamily protein] |
-0.350 |
1.025 |
| 452 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.349 |
0.318 |
| 453 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
-0.348 |
0.015 |
| 454 |
253403_at |
AT4G32830
|
AtAUR1, ataurora1, AUR1, ataurora1 |
-0.347 |
0.643 |
| 455 |
253578_at |
AT4G30340
|
ATDGK7, diacylglycerol kinase 7, DGK7, diacylglycerol kinase 7 |
-0.346 |
0.739 |
| 456 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.346 |
0.915 |
| 457 |
257020_at |
AT3G19590
|
BUB3.1, BUB (BUDDING UNINHIBITED BY BENZYMIDAZOL) 3.1 |
-0.346 |
0.387 |
| 458 |
260656_at |
AT1G19380
|
unknown |
-0.345 |
-0.800 |
| 459 |
265695_at |
AT2G24490
|
ATRPA2, ATRPA32A, ROR1, SUPPRESSOR OF ROS1, RPA2, replicon protein A2, RPA32A |
-0.344 |
1.632 |
| 460 |
251284_at |
AT3G61840
|
unknown |
-0.344 |
-0.852 |
| 461 |
254226_at |
AT4G23690
|
AtDIR6, Arabidopsis thaliana dirigent protein 6, DIR6, dirigent protein 6 |
-0.342 |
-0.437 |
| 462 |
266489_at |
AT2G35190
|
ATNPSN11, NPSN11, novel plant snare 11, NSPN11 |
-0.340 |
0.990 |
| 463 |
258002_at |
AT3G28930
|
AIG2, AVRRPT2-INDUCED GENE 2 |
-0.340 |
-0.651 |
| 464 |
247358_at |
AT5G63580
|
ATFLS2, FLS2, flavonol synthase 2 |
-0.339 |
-0.529 |
| 465 |
253389_at |
AT4G32680
|
unknown |
-0.339 |
0.118 |
| 466 |
256157_at |
AT1G13580
|
LAG13, LAG1 longevity assurance homolog 3, LOH3, LAG One Homologue 3 |
-0.338 |
0.244 |
| 467 |
251370_at |
AT3G60450
|
[Phosphoglycerate mutase family protein] |
-0.338 |
0.082 |
| 468 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
-0.337 |
-0.765 |
| 469 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
-0.337 |
-0.325 |
| 470 |
257134_at |
AT3G12870
|
unknown |
-0.335 |
0.661 |
| 471 |
262919_at |
AT1G79380
|
RGLG4, RING DOMAIN LIGASE 4 |
-0.335 |
0.965 |
| 472 |
263797_at |
AT2G24570
|
ATWRKY17, WRKY17, WRKY DNA-binding protein 17 |
-0.334 |
1.102 |
| 473 |
250337_at |
AT5G11790
|
NDL2, N-MYC downregulated-like 2 |
-0.333 |
-0.109 |
| 474 |
260081_at |
AT1G78170
|
unknown |
-0.333 |
0.338 |
| 475 |
251661_at |
AT3G56950
|
SIP2, SMALL AND BASIC INTRINSIC PROTEIN 2, SIP2;1, small and basic intrinsic protein 2;1 |
-0.333 |
0.701 |
| 476 |
245612_at |
AT4G14440
|
ATECI3, ARABIDOPSIS THALIANA DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, ECI3, DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, HCD1, 3-hydroxyacyl-CoA dehydratase 1 |
-0.333 |
0.904 |
| 477 |
247284_at |
AT5G64410
|
ATOPT4, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 4, OPT4, oligopeptide transporter 4 |
-0.332 |
-0.225 |
| 478 |
263714_at |
AT2G20610
|
ALF1, ABERRANT LATERAL ROOT FORMATION 1, HLS3, HOOKLESS 3, RTY, ROOTY, RTY1, ROOTY 1, SUR1, SUPERROOT 1 |
-0.332 |
0.132 |
| 479 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
-0.331 |
-0.723 |
| 480 |
266811_at |
AT2G44850
|
unknown |
-0.331 |
-0.828 |
| 481 |
252821_at |
AT4G39860
|
unknown |
-0.331 |
1.114 |
| 482 |
265656_at |
AT2G13820
|
AtXYP2, XYP2, xylogen protein 2 |
-0.331 |
0.589 |
| 483 |
265075_at |
AT1G55450
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.330 |
-0.555 |
| 484 |
255460_at |
AT4G02800
|
unknown |
-0.329 |
1.255 |
| 485 |
264343_at |
AT1G11850
|
unknown |
-0.329 |
0.706 |
| 486 |
263619_at |
AT2G04650
|
[ADP-glucose pyrophosphorylase family protein] |
-0.329 |
0.017 |
| 487 |
265539_at |
AT2G15830
|
unknown |
-0.329 |
-0.497 |
| 488 |
260074_at |
AT1G73640
|
AtRABA6a, RAB GTPase homolog A6A, RABA6a, RAB GTPase homolog A6A |
-0.329 |
-0.341 |
| 489 |
264998_at |
AT1G67330
|
unknown |
-0.329 |
-0.292 |
| 490 |
253122_at |
AT4G35987
|
CaM KMT, calmodulin N-methyltransferase |
-0.328 |
-0.479 |
| 491 |
261525_at |
AT1G14330
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.328 |
0.319 |
| 492 |
259160_at |
AT3G05410
|
[Photosystem II reaction center PsbP family protein] |
-0.328 |
-0.389 |
| 493 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
-0.327 |
-0.676 |
| 494 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
-0.326 |
-0.859 |
| 495 |
258040_at |
AT3G21190
|
AtMSR1, MSR1, Mannan Synthesis Related 1 |
-0.326 |
-0.173 |
| 496 |
267165_at |
AT2G37710
|
LecRK-IV.1, L-type lectin receptor kinase IV.1, RLK, receptor lectin kinase |
-0.324 |
0.812 |
| 497 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
-0.322 |
0.407 |
| 498 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-0.322 |
-0.547 |
| 499 |
247255_at |
AT5G64780
|
[Uncharacterised conserved protein UCP009193] |
-0.321 |
-0.763 |
| 500 |
248082_at |
AT5G55400
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.319 |
-0.682 |
| 501 |
250490_at |
AT5G09760
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.319 |
0.214 |
| 502 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
-0.318 |
0.389 |
| 503 |
246517_at |
AT5G15760
|
PSRP3/2, plastid-specific ribosomal protein 3/2 |
-0.318 |
0.819 |
| 504 |
253401_at |
AT4G32870
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.317 |
-0.274 |
| 505 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.317 |
0.400 |
| 506 |
250745_at |
AT5G05850
|
PIRL1, plant intracellular ras group-related LRR 1 |
-0.317 |
-0.640 |
| 507 |
252504_at |
AT3G46590
|
ATTRP2, TRFL1, TRF-like 1, TRP2 |
-0.316 |
-0.113 |
| 508 |
264321_at |
AT1G04200
|
unknown |
-0.312 |
-0.338 |
| 509 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
-0.311 |
0.544 |
| 510 |
255031_at |
AT4G09490
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
-0.311 |
-0.744 |
| 511 |
249096_at |
AT5G43910
|
[pfkB-like carbohydrate kinase family protein] |
-0.309 |
-1.008 |
| 512 |
265994_at |
AT2G24170
|
[Endomembrane protein 70 protein family] |
-0.309 |
0.213 |
| 513 |
250580_at |
AT5G07440
|
GDH2, glutamate dehydrogenase 2 |
-0.309 |
0.457 |
| 514 |
258067_at |
AT3G25980
|
AtMAD2, MAD2, MITOTIC ARREST-DEFICIENT 2 |
-0.309 |
-0.080 |
| 515 |
258782_at |
AT3G11750
|
FOLB1 |
-0.308 |
-1.187 |
| 516 |
266770_at |
AT2G03090
|
ATEXP15, ATEXPA15, expansin A15, ATHEXP ALPHA 1.3, EXP15, EXPANSIN 15, EXPA15, expansin A15 |
-0.307 |
0.165 |
| 517 |
245414_at |
AT4G17310
|
unknown |
-0.306 |
0.303 |
| 518 |
262772_at |
AT1G13210
|
ACA.l, autoinhibited Ca2+/ATPase II |
-0.306 |
0.725 |
| 519 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
-0.306 |
-0.153 |
| 520 |
248772_at |
AT5G47800
|
[Phototropic-responsive NPH3 family protein] |
-0.306 |
-0.360 |
| 521 |
248006_at |
AT5G56250
|
HAP8, HAPLESS 8 |
-0.305 |
-0.157 |
| 522 |
252859_at |
AT4G39780
|
[Integrase-type DNA-binding superfamily protein] |
-0.305 |
0.011 |
| 523 |
257553_at |
AT3G16830
|
TPR2, TOPLESS-related 2 |
-0.304 |
0.423 |
| 524 |
252121_at |
AT3G51160
|
GMD2, GDP-D-MANNOSE-4,6-DEHYDRATASE 2, MUR1, MURUS 1, MUR_1, MURUS 1 |
-0.304 |
1.002 |
| 525 |
262566_at |
AT1G34310
|
ARF12, auxin response factor 12 |
-0.303 |
0.414 |
| 526 |
246135_at |
AT5G20885
|
[RING/U-box superfamily protein] |
-0.303 |
-0.025 |
| 527 |
255561_at |
AT4G02050
|
STP7, sugar transporter protein 7 |
-0.303 |
0.560 |
| 528 |
254229_at |
AT4G23610
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.303 |
-0.357 |
| 529 |
248756_at |
AT5G47560
|
ATSDAT, ATTDT, TONOPLAST DICARBOXYLATE TRANSPORTER, TDT, tonoplast dicarboxylate transporter |
-0.303 |
0.146 |
| 530 |
261921_at |
AT1G65900
|
unknown |
-0.302 |
1.283 |
| 531 |
260028_at |
AT1G29980
|
unknown |
-0.302 |
0.371 |
| 532 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.300 |
0.588 |
| 533 |
267209_at |
AT2G30930
|
unknown |
-0.300 |
-0.540 |
| 534 |
262408_at |
AT1G34750
|
[Protein phosphatase 2C family protein] |
-0.299 |
0.291 |
| 535 |
249078_at |
AT5G44070
|
ARA8, ATPCS1, ARABIDOPSIS THALIANA PHYTOCHELATIN SYNTHASE 1, CAD1, CADMIUM SENSITIVE 1, PCS1, PHYTOCHELATIN SYNTHASE 1 |
-0.299 |
0.360 |
| 536 |
263628_at |
AT2G04780
|
FLA7, FASCICLIN-like arabinoogalactan 7 |
-0.297 |
0.224 |
| 537 |
262134_at |
AT1G77990
|
AST56, SULTR2;2, SULPHATE TRANSPORTER 2;2 |
-0.297 |
-0.878 |
| 538 |
256529_at |
AT1G33260
|
[Protein kinase superfamily protein] |
-0.296 |
-1.298 |
| 539 |
246144_at |
AT5G20110
|
[Dynein light chain type 1 family protein] |
-0.296 |
-0.674 |
| 540 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.295 |
0.596 |
| 541 |
262414_at |
AT1G49430
|
LACS2, long-chain acyl-CoA synthetase 2, LRD2, LATERAL ROOT DEVELOPMENT 2 |
-0.293 |
0.729 |
| 542 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
-0.293 |
-0.420 |
| 543 |
256469_at |
AT1G32540
|
LOL1, lsd one like 1 |
-0.292 |
1.057 |
| 544 |
261605_at |
AT1G49580
|
[Calcium-dependent protein kinase (CDPK) family protein] |
-0.292 |
0.224 |
| 545 |
261826_at |
AT1G11580
|
ATPMEPCRA, PMEPCRA, methylesterase PCR A |
-0.290 |
0.215 |
| 546 |
266802_at |
AT2G22900
|
[Galactosyl transferase GMA12/MNN10 family protein] |
-0.290 |
0.340 |
| 547 |
263934_at |
AT2G35950
|
EDA12, embryo sac development arrest 12 |
-0.289 |
-0.831 |
| 548 |
248269_at |
AT5G53470
|
ACBP, ACYL-COA BINDING PROTEIN, ACBP1, acyl-CoA binding protein 1, AtACBP1 |
-0.289 |
-0.194 |
| 549 |
250743_at |
AT5G05820
|
[Nucleotide-sugar transporter family protein] |
-0.288 |
0.869 |
| 550 |
261455_at |
AT1G21070
|
[Nucleotide-sugar transporter family protein] |
-0.287 |
-0.287 |
| 551 |
261070_at |
AT1G07390
|
AtRLP1, receptor like protein 1, RLP1, receptor like protein 1 |
-0.285 |
-0.068 |
| 552 |
263838_at |
AT2G36880
|
MAT3, methionine adenosyltransferase 3 |
-0.285 |
1.577 |
| 553 |
257939_at |
AT3G19930
|
ATSTP4, SUGAR TRANSPORTER 4, STP4, sugar transporter 4 |
-0.285 |
0.596 |
| 554 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
-0.284 |
-0.261 |
| 555 |
258049_at |
AT3G16220
|
[Predicted eukaryotic LigT] |
-0.284 |
-0.640 |
| 556 |
256681_at |
AT3G52340
|
ATSPP2, SUCROSE-PHOSPHATASE 2, SPP2, sucrose-6F-phosphate phosphohydrolase 2 |
-0.284 |
0.453 |
| 557 |
245739_at |
AT1G44110
|
CYCA1;1, Cyclin A1;1 |
-0.283 |
0.994 |
| 558 |
259721_at |
AT1G60890
|
[Phosphatidylinositol-4-phosphate 5-kinase family protein] |
-0.282 |
0.348 |
| 559 |
260226_at |
AT1G74660
|
MIF1, mini zinc finger 1 |
-0.281 |
-0.971 |
| 560 |
263570_at |
AT2G27150
|
AAO3, abscisic aldehyde oxidase 3, AOdelta, Aldehyde oxidase delta, At-AO3, Arabidopsis thaliana aldehyde oxidase 3, AtAAO3 |
-0.281 |
-0.831 |
| 561 |
262208_at |
AT1G74800
|
[Galactosyltransferase family protein] |
-0.280 |
1.193 |
| 562 |
260002_at |
AT1G67940
|
ABCI17, ATP-binding cassette I17, ATNAP3, non-intrinsic ABC protein 3, AtSTAR1, NAP3, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 3, NAP3, non-intrinsic ABC protein 3 |
-0.280 |
-0.014 |
| 563 |
260167_at |
AT1G71970
|
unknown |
-0.279 |
1.010 |
| 564 |
255637_at |
AT4G00750
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.279 |
-0.500 |
| 565 |
256150_at |
AT1G55120
|
AtcwINV3, 6-fructan exohydrolase, ATFRUCT5, beta-fructofuranosidase 5, FRUCT5, beta-fructofuranosidase 5 |
-0.279 |
0.136 |
| 566 |
255628_at |
AT4G00850
|
GIF3, GRF1-interacting factor 3 |
-0.279 |
0.642 |
| 567 |
257466_at |
AT1G62840
|
unknown |
-0.278 |
-0.756 |
| 568 |
262694_at |
AT1G62790
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.278 |
0.814 |
| 569 |
251048_at |
AT5G02410
|
ALG10, homolog of yeast ALG10 |
-0.277 |
0.994 |
| 570 |
246505_at |
AT5G16250
|
unknown |
-0.277 |
0.812 |
| 571 |
263618_at |
AT2G04660
|
APC2, anaphase-promoting complex/cyclosome 2 |
-0.275 |
-0.352 |
| 572 |
255543_at |
AT4G01870
|
[tolB protein-related] |
-0.275 |
0.481 |
| 573 |
263151_at |
AT1G54120
|
unknown |
-0.274 |
-0.588 |
| 574 |
245197_at |
AT1G67800
|
[Copine (Calcium-dependent phospholipid-binding protein) family] |
-0.273 |
0.079 |
| 575 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
-0.273 |
1.367 |
| 576 |
258953_at |
AT3G01430
|
unknown |
-0.272 |
0.190 |
| 577 |
264852_at |
AT2G17480
|
ATMLO8, MILDEW RESISTANCE LOCUS O 8, MLO8, MILDEW RESISTANCE LOCUS O 8 |
-0.272 |
0.219 |
| 578 |
264669_at |
AT1G09630
|
ATRAB-A2A, ARABIDOPSIS RAB GTPASE A2A, ATRAB11C, RAB GTPase 11C, ATRABA2A, ARABIDOPSIS RAB GTPASE A2A, RAB-A2A, RAB GTPASE A2A, RAB11c, RAB GTPase 11C |
-0.271 |
0.665 |
| 579 |
245740_at |
AT1G44100
|
AAP5, amino acid permease 5 |
-0.271 |
-0.027 |
| 580 |
248008_at |
AT5G56270
|
ATWRKY2, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 2, WRKY2, WRKY DNA-binding protein 2 |
-0.271 |
-0.426 |
| 581 |
253500_at |
AT4G31920
|
ARR10, response regulator 10, RR10, response regulator 10 |
-0.271 |
-0.223 |
| 582 |
264379_at |
AT2G25200
|
unknown |
-0.271 |
-0.825 |
| 583 |
248769_at |
AT5G47730
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.270 |
0.109 |
| 584 |
247643_at |
AT5G60450
|
ARF4, auxin response factor 4 |
-0.270 |
0.969 |
| 585 |
261791_at |
AT1G16170
|
unknown |
-0.270 |
-0.203 |
| 586 |
261470_at |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
-0.270 |
-0.432 |
| 587 |
259980_at |
AT1G76520
|
[Auxin efflux carrier family protein] |
-0.270 |
-0.372 |
| 588 |
264668_at |
AT1G09780
|
iPGAM1, 2,3-biphosphoglycerate-independent phosphoglycerate mutase 1 |
-0.269 |
1.501 |
| 589 |
249411_at |
AT5G40390
|
RS5, raffinose synthase 5, SIP1, seed imbibition 1-like |
-0.269 |
-0.217 |
| 590 |
259987_at |
AT1G75030
|
ATLP-3, thaumatin-like protein 3, TLP-3, thaumatin-like protein 3 |
-0.268 |
-0.042 |
| 591 |
253753_at |
AT4G29030
|
[Putative membrane lipoprotein] |
-0.268 |
0.450 |
| 592 |
254284_at |
AT4G22910
|
CCS52A1, cell cycle switch protein 52 A1, FZR2, FIZZY-related 2 |
-0.268 |
-0.040 |
| 593 |
252057_at |
AT3G52480
|
unknown |
-0.268 |
-0.753 |
| 594 |
252624_at |
AT3G44735
|
ATPSK3, PHYTOSULFOKINE 3 PRECURSOR, PSK1, PSK3, PHYTOSULFOKINE 3 PRECURSOR |
-0.267 |
0.473 |
| 595 |
247030_at |
AT5G67210
|
IRX15-L, IRX15-LIKE |
-0.266 |
0.730 |
| 596 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
-0.266 |
-0.684 |
| 597 |
248623_at |
AT5G49170
|
unknown |
-0.266 |
0.773 |
| 598 |
263913_at |
AT2G36570
|
PXC1, PXY/TDR-correlated 1 |
-0.266 |
0.879 |
| 599 |
255860_at |
AT5G34940
|
AtGUS3, glucuronidase 3, GUS3, glucuronidase 3 |
-0.266 |
0.542 |
| 600 |
254573_at |
AT4G19420
|
[Pectinacetylesterase family protein] |
-0.266 |
-0.397 |