|
probeID |
AGICode |
Annotation |
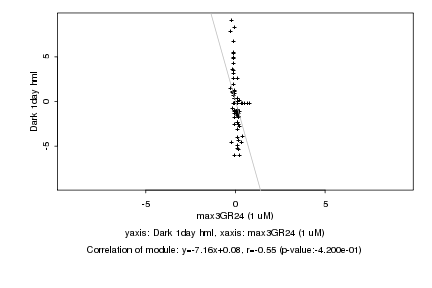
Log2 signal ratio
max3GR24 (1 uM) |
Log2 signal ratio
Dark 1day hml |
| 1 |
265891_at |
AT2G15010
|
[Plant thionin] |
0.755 |
-0.175 |
| 2 |
250468_at |
AT5G10120
|
[Ethylene insensitive 3 family protein] |
0.629 |
-0.175 |
| 3 |
262431_at |
AT1G47540
|
[Scorpion toxin-like knottin superfamily protein] |
0.472 |
-0.175 |
| 4 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.393 |
-3.861 |
| 5 |
250610_at |
AT5G07550
|
ATGRP19, GRP19, glycine-rich protein 19 |
0.351 |
-0.175 |
| 6 |
246001_at |
AT5G20790
|
unknown |
0.314 |
-4.529 |
| 7 |
249848_at |
AT5G23220
|
NIC3, nicotinamidase 3 |
0.289 |
-0.175 |
| 8 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
0.213 |
0.199 |
| 9 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.204 |
-5.998 |
| 10 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
0.202 |
-2.685 |
| 11 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.187 |
-1.096 |
| 12 |
260363_at |
AT1G70550
|
unknown |
0.163 |
-1.658 |
| 13 |
257899_at |
AT3G28345
|
ABCB15, ATP-binding cassette B15, MDR13, multi-drug resistance 13 |
0.158 |
-4.293 |
| 14 |
260053_at |
AT1G78120
|
TPR12, tetratricopeptide repeat 12 |
0.156 |
-2.507 |
| 15 |
247657_at |
AT5G59845
|
[Gibberellin-regulated family protein] |
0.138 |
-1.341 |
| 16 |
264527_at |
AT1G30760
|
[FAD-binding Berberine family protein] |
0.133 |
-1.653 |
| 17 |
262081_at |
AT1G59540
|
ZCF125 |
0.128 |
-1.708 |
| 18 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
0.123 |
-1.019 |
| 19 |
263441_at |
AT2G28620
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.120 |
-5.276 |
| 20 |
258480_at |
AT3G02640
|
unknown |
0.114 |
-3.940 |
| 21 |
263599_at |
AT2G01830
|
AHK4, ARABIDOPSIS HISTIDINE KINASE 4, ATCRE1, CRE1, CYTOKININ RESPONSE 1, WOL, WOODEN LEG, WOL1, WOODEN LEG 1 |
0.112 |
-1.064 |
| 22 |
259897_at |
AT1G71380
|
ATCEL3, cellulase 3, ATGH9B3, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B3, CEL3, cellulase 3 |
0.111 |
-0.175 |
| 23 |
255852_at |
AT1G66970
|
GDPDL1, Glycerophosphodiester phosphodiesterase (GDPD) like 1, SVL2, SHV3-like 2 |
0.110 |
-1.320 |
| 24 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
0.109 |
-0.969 |
| 25 |
264998_at |
AT1G67330
|
unknown |
0.104 |
-2.321 |
| 26 |
265339_at |
AT2G18230
|
AtPPa2, pyrophosphorylase 2, PPa2, pyrophosphorylase 2 |
0.101 |
-1.527 |
| 27 |
254419_at |
AT4G21490
|
NDB3, NAD(P)H dehydrogenase B3 |
0.100 |
0.352 |
| 28 |
248406_at |
AT5G51490
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.098 |
-0.175 |
| 29 |
251419_at |
AT3G60470
|
unknown |
0.097 |
-4.867 |
| 30 |
265188_at |
AT1G23800
|
ALDH2B, aldehyde dehydrogenase 2B, ALDH2B7, aldehyde dehydrogenase 2B7 |
0.097 |
-1.158 |
| 31 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.096 |
-1.131 |
| 32 |
262198_at |
AT1G53830
|
ATPME2, pectin methylesterase 2, PME2, pectin methylesterase 2 |
0.094 |
0.104 |
| 33 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.093 |
-0.805 |
| 34 |
249214_at |
AT5G42720
|
[Glycosyl hydrolase family 17 protein] |
0.093 |
-3.033 |
| 35 |
255590_at |
AT4G01610
|
[Cysteine proteinases superfamily protein] |
0.092 |
2.605 |
| 36 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
0.090 |
-5.258 |
| 37 |
262558_at |
AT1G31335
|
unknown |
0.089 |
-1.134 |
| 38 |
253758_at |
AT4G29060
|
emb2726, embryo defective 2726 |
0.088 |
-1.403 |
| 39 |
267382_at |
AT2G44300
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.088 |
-2.253 |
| 40 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
-0.323 |
7.944 |
| 41 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
-0.303 |
1.500 |
| 42 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
-0.274 |
-4.548 |
| 43 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
-0.255 |
9.182 |
| 44 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
-0.210 |
1.069 |
| 45 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
-0.203 |
-0.709 |
| 46 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
-0.200 |
1.148 |
| 47 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
-0.189 |
3.645 |
| 48 |
263118_at |
AT1G03090
|
MCCA |
-0.154 |
5.585 |
| 49 |
260930_at |
AT1G02620
|
[Ras-related small GTP-binding family protein] |
-0.145 |
5.421 |
| 50 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.140 |
3.476 |
| 51 |
250629_at |
AT5G07390
|
ATRBOHA, respiratory burst oxidase homolog A, RBOHA, respiratory burst oxidase homolog A |
-0.140 |
0.738 |
| 52 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
-0.139 |
-0.203 |
| 53 |
267391_at |
AT2G44480
|
BGLU17, beta glucosidase 17 |
-0.134 |
1.974 |
| 54 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
-0.132 |
2.590 |
| 55 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
-0.131 |
4.898 |
| 56 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
-0.129 |
4.942 |
| 57 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
-0.129 |
4.350 |
| 58 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.128 |
0.908 |
| 59 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.124 |
6.775 |
| 60 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.116 |
3.237 |
| 61 |
251491_at |
AT3G59480
|
[pfkB-like carbohydrate kinase family protein] |
-0.108 |
-1.690 |
| 62 |
266733_at |
AT2G03280
|
[O-fucosyltransferase family protein] |
-0.107 |
-1.043 |
| 63 |
257421_at |
AT1G12030
|
unknown |
-0.105 |
-2.538 |
| 64 |
259500_at |
AT1G15740
|
[Leucine-rich repeat family protein] |
-0.104 |
1.217 |
| 65 |
246049_at |
AT5G28890
|
[similar to DNA binding [Arabidopsis thaliana] (TAIR:AT4G01980.1)] |
-0.104 |
-0.175 |
| 66 |
255107_at |
AT4G08730
|
[RNA-binding protein] |
-0.104 |
-0.175 |
| 67 |
252451_at |
AT3G47100
|
unknown |
-0.103 |
-0.915 |
| 68 |
249566_at |
AT5G38450
|
CYP735A1, cytochrome P450, family 735, subfamily A, polypeptide 1 |
-0.101 |
-0.175 |
| 69 |
248870_at |
AT5G46710
|
[PLATZ transcription factor family protein] |
-0.101 |
1.319 |
| 70 |
247195_at |
AT5G65500
|
[U-box domain-containing protein kinase family protein] |
-0.099 |
-1.336 |
| 71 |
258342_at |
AT3G22800
|
[Leucine-rich repeat (LRR) family protein] |
-0.098 |
-0.175 |
| 72 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
-0.097 |
8.358 |
| 73 |
246208_at |
AT4G36490
|
ATSFH12, SEC14-LIKE 12, SFH12, SEC14-like 12 |
-0.095 |
-0.175 |
| 74 |
253195_at |
AT4G35420
|
DRL1, dihydroflavonol 4-reductase-like1, TKPR1, tetraketide alpha-pyrone reductase 1 |
-0.095 |
0.903 |
| 75 |
257772_at |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.095 |
0.414 |
| 76 |
259650_at |
AT1G55290
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
-0.094 |
0.352 |
| 77 |
261712_at |
AT1G32780
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.094 |
-6.028 |