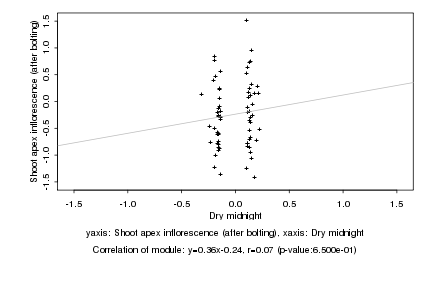
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Dry midnight |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
256603_at |
AT3G28270
|
unknown |
0.218 |
-0.506 |
| 2 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
0.213 |
0.157 |
| 3 |
260576_at |
AT2G47310
|
[flowering time control protein-related / FCA gamma-related] |
0.197 |
0.296 |
| 4 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
0.190 |
-0.723 |
| 5 |
254548_at |
AT4G19865
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.171 |
0.168 |
| 6 |
258935_at |
AT3G10120
|
unknown |
0.171 |
-1.408 |
| 7 |
259001_at |
AT3G01960
|
unknown |
0.157 |
-0.260 |
| 8 |
259987_at |
AT1G75030
|
ATLP-3, thaumatin-like protein 3, TLP-3, thaumatin-like protein 3 |
0.151 |
-0.042 |
| 9 |
247913_at |
AT5G57510
|
unknown |
0.144 |
-1.049 |
| 10 |
249792_at |
AT5G23720
|
PHS1, PROPYZAMIDE-HYPERSENSITIVE 1 |
0.142 |
0.954 |
| 11 |
265926_at |
AT2G18600
|
[Ubiquitin-conjugating enzyme family protein] |
0.140 |
0.329 |
| 12 |
265078_at |
AT1G55500
|
ECT4, evolutionarily conserved C-terminal region 4 |
0.137 |
-0.297 |
| 13 |
262465_at |
AT1G50270
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.136 |
0.128 |
| 14 |
266525_at |
AT2G16970
|
MEE15, maternal effect embryo arrest 15 |
0.134 |
-0.378 |
| 15 |
263974_at |
AT2G42720
|
[FBD, F-box, Skp2-like and Leucine Rich Repeat domains containing protein] |
0.133 |
-0.656 |
| 16 |
261219_at |
AT1G19980
|
[cytomatrix protein-related] |
0.132 |
0.757 |
| 17 |
251699_at |
AT3G56630
|
CYP94D2, cytochrome P450, family 94, subfamily D, polypeptide 2 |
0.132 |
-0.946 |
| 18 |
251752_at |
AT3G55740
|
ATPROT2, PROLINE TRANSPORTER 2, PROT2, proline transporter 2 |
0.128 |
-0.853 |
| 19 |
246685_at |
AT5G33350
|
[pseudogene] |
0.127 |
-0.170 |
| 20 |
259945_at |
AT1G71460
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.124 |
0.746 |
| 21 |
257374_at |
AT2G43280
|
[Far-red impaired responsive (FAR1) family protein] |
0.124 |
-0.349 |
| 22 |
262777_at |
AT1G13030
|
Atcoilin, COILIN, coilin |
0.124 |
0.256 |
| 23 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
0.123 |
-0.702 |
| 24 |
252767_at |
AT3G42830
|
[RING/U-box superfamily protein] |
0.122 |
-0.523 |
| 25 |
259142_at |
AT3G10200
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.119 |
0.082 |
| 26 |
266362_at |
AT2G32430
|
[Galactosyltransferase family protein] |
0.115 |
0.182 |
| 27 |
260264_at |
AT1G68500
|
unknown |
0.112 |
-0.773 |
| 28 |
260939_at |
AT1G45180
|
[RING/U-box superfamily protein] |
0.111 |
0.637 |
| 29 |
267646_at |
AT2G32830
|
PHT1;5, phosphate transporter 1;5, PHT5, PHOSPHATE TRANSPORTER 5 |
0.110 |
-0.839 |
| 30 |
251955_at |
AT3G53680
|
[Acyl-CoA N-acyltransferase with RING/FYVE/PHD-type zinc finger domain] |
0.107 |
-0.775 |
| 31 |
246557_at |
AT5G15510
|
[TPX2 (targeting protein for Xklp2) protein family] |
0.105 |
-0.094 |
| 32 |
258260_at |
AT3G26850
|
[histone-lysine N-methyltransferases] |
0.104 |
-0.192 |
| 33 |
246598_at |
AT5G14850
|
[Alg9-like mannosyltransferase family] |
0.101 |
0.532 |
| 34 |
264286_at |
AT1G61870
|
PPR336, pentatricopeptide repeat 336 |
0.101 |
1.530 |
| 35 |
265405_at |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
0.098 |
-1.237 |
| 36 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
-0.322 |
0.138 |
| 37 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
-0.243 |
-0.452 |
| 38 |
258008_at |
AT3G19430
|
[late embryogenesis abundant protein-related / LEA protein-related] |
-0.239 |
-0.755 |
| 39 |
264635_at |
AT1G65500
|
unknown |
-0.208 |
0.393 |
| 40 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.201 |
0.773 |
| 41 |
248609_at |
AT5G49440
|
unknown |
-0.199 |
-0.499 |
| 42 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.196 |
0.844 |
| 43 |
251201_at |
AT3G63020
|
unknown |
-0.195 |
-1.213 |
| 44 |
252780_at |
AT3G42960
|
ASD, ATA1, TAPETUM 1, TA1, TAPETUM1 |
-0.194 |
-1.006 |
| 45 |
255543_at |
AT4G01870
|
[tolB protein-related] |
-0.186 |
0.481 |
| 46 |
263013_at |
AT1G23380
|
KNAT6, KNOTTED1-like homeobox gene 6, KNAT6L, KNAT6S |
-0.174 |
-0.194 |
| 47 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
-0.174 |
-0.600 |
| 48 |
256716_at |
AT2G34100
|
unknown |
-0.168 |
-0.773 |
| 49 |
245640_at |
AT1G25330
|
CES, CESTA, HAF, HALF FILLED |
-0.167 |
-0.565 |
| 50 |
245802_at |
AT1G46840
|
[F-box family protein] |
-0.166 |
-0.902 |
| 51 |
264538_at |
AT1G55600
|
ATWRKY10, MINI3, MINISEED 3, WRKY10, WRKY DNA-binding protein 10 |
-0.165 |
-0.598 |
| 52 |
263947_at |
AT2G35820
|
[ureidoglycolate hydrolases] |
-0.164 |
-0.745 |
| 53 |
254718_at |
AT4G13580
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.163 |
-0.254 |
| 54 |
267493_at |
AT2G30400
|
ATOFP2, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 2, OFP2, ovate family protein 2 |
-0.162 |
-0.791 |
| 55 |
261262_at |
AT1G26760
|
ATXR1, SDG35, SET domain protein 35 |
-0.161 |
-0.850 |
| 56 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.159 |
-0.582 |
| 57 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
-0.159 |
-0.114 |
| 58 |
260642_at |
AT1G53260
|
[LOCATED IN: endomembrane system] |
-0.159 |
-0.277 |
| 59 |
267620_at |
AT2G39640
|
[glycosyl hydrolase family 17 protein] |
-0.157 |
0.062 |
| 60 |
264830_at |
AT1G03710
|
[Cystatin/monellin superfamily protein] |
-0.156 |
0.251 |
| 61 |
247895_at |
AT5G58010
|
LRL3, LJRHL1-like 3 |
-0.154 |
0.226 |
| 62 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.151 |
-0.872 |
| 63 |
250169_at |
AT5G15340
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.149 |
-0.085 |
| 64 |
247764_at |
AT5G59190
|
[subtilase family protein] |
-0.148 |
-0.271 |
| 65 |
265115_at |
AT1G62450
|
[Immunoglobulin E-set superfamily protein] |
-0.148 |
-0.175 |
| 66 |
263427_at |
AT2G22260
|
ALKBH2, homolog of E. coli alkB |
-0.146 |
0.573 |
| 67 |
255518_at |
AT4G02300
|
PME39, pectin methylesterase 39 |
-0.146 |
-0.177 |
| 68 |
261245_at |
AT1G20135
|
[GDSL-like Lipase/Acylhydrolase family protein] |
-0.145 |
-1.359 |
| 69 |
253618_at |
AT4G30420
|
UMAMIT34, Usually multiple acids move in and out Transporters 34 |
-0.145 |
-0.319 |
| 70 |
250415_at |
AT5G11210
|
ATGLR2.5, ARABIDOPSIS THALIANA GLU, GLR2.5, glutamate receptor 2.5 |
-0.145 |
-0.297 |