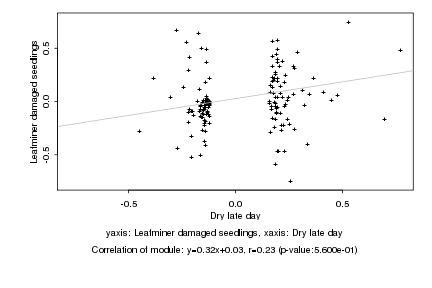
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Dry late day |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.770 |
0.487 |
| 2 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
0.696 |
-0.168 |
| 3 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.528 |
0.749 |
| 4 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.474 |
0.065 |
| 5 |
256924_at |
AT3G29590
|
AT5MAT |
0.449 |
0.013 |
| 6 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
0.410 |
0.086 |
| 7 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
0.386 |
0.964 |
| 8 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.363 |
0.224 |
| 9 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.346 |
0.073 |
| 10 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.334 |
-0.402 |
| 11 |
247751_at |
AT5G59050
|
unknown |
0.321 |
-0.032 |
| 12 |
265913_at |
AT2G25625
|
unknown |
0.313 |
0.111 |
| 13 |
257925_at |
AT3G23170
|
unknown |
0.289 |
0.469 |
| 14 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.274 |
0.315 |
| 15 |
246468_at |
AT5G17050
|
UGT78D2, UDP-glucosyl transferase 78D2 |
0.274 |
-0.258 |
| 16 |
251644_at |
AT3G57540
|
[Remorin family protein] |
0.270 |
0.338 |
| 17 |
253219_at |
AT4G34990
|
AtMYB32, myb domain protein 32, MYB32, myb domain protein 32 |
0.270 |
0.075 |
| 18 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.255 |
-0.745 |
| 19 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
0.250 |
-0.213 |
| 20 |
266488_at |
AT2G47670
|
PMEI6, PECTIN METHYLESTERASE INHIBITOR 6 |
0.246 |
0.044 |
| 21 |
258962_at |
AT3G10570
|
CYP77A6, cytochrome P450, family 77, subfamily A, polypeptide 6 |
0.242 |
-0.168 |
| 22 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.240 |
1.262 |
| 23 |
249601_at |
AT5G37980
|
[Zinc-binding dehydrogenase family protein] |
0.239 |
0.011 |
| 24 |
265359_at |
AT2G16720
|
ATMYB7, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 7, ATY49, MYB7, myb domain protein 7 |
0.237 |
0.869 |
| 25 |
266617_at |
AT2G29670
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.234 |
0.250 |
| 26 |
256603_at |
AT3G28270
|
unknown |
0.233 |
-0.020 |
| 27 |
261785_at |
AT1G08230
|
[Transmembrane amino acid transporter family protein] |
0.229 |
0.182 |
| 28 |
258932_at |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
0.227 |
-0.461 |
| 29 |
260851_at |
AT1G21890
|
UMAMIT19, Usually multiple acids move in and out Transporters 19 |
0.226 |
-0.041 |
| 30 |
252888_at |
AT4G39210
|
APL3 |
0.224 |
-0.222 |
| 31 |
258362_at |
AT3G14280
|
unknown |
0.220 |
0.383 |
| 32 |
254186_at |
AT4G24010
|
ATCSLG1, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G1, CSLG1, cellulose synthase like G1 |
0.218 |
0.040 |
| 33 |
264692_at |
AT1G70000
|
[myb-like transcription factor family protein] |
0.213 |
-0.218 |
| 34 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.211 |
-0.268 |
| 35 |
259970_at |
AT1G76570
|
AtLHCB7, LHCB7, light-harvesting complex B7 |
0.210 |
-0.109 |
| 36 |
262803_at |
AT1G21000
|
[PLATZ transcription factor family protein] |
0.207 |
0.077 |
| 37 |
260156_at |
AT1G52880
|
ANAC018, Arabidopsis NAC domain containing protein 18, ATNAM, NAM, NO APICAL MERISTEM, NARS2, NAC-REGULATED SEED MORPHOLOGY 2 |
0.207 |
0.144 |
| 38 |
255430_at |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
0.203 |
0.329 |
| 39 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
0.201 |
-0.466 |
| 40 |
264580_at |
AT1G05340
|
unknown |
0.200 |
0.981 |
| 41 |
262073_at |
AT1G59640
|
BPE, BIG PETAL, BPEp, BIG PETAL P, BPEub, BIG PETAL UB, ZCW32 |
0.196 |
0.193 |
| 42 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
0.196 |
-0.052 |
| 43 |
254707_at |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
0.195 |
0.370 |
| 44 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
0.195 |
0.581 |
| 45 |
245348_at |
AT4G17770
|
ATTPS5, trehalose phosphatase/synthase 5, TPS5, TREHALOSE -6-PHOSPHATASE SYNTHASE S5, TPS5, trehalose phosphatase/synthase 5 |
0.195 |
-0.466 |
| 46 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.194 |
0.398 |
| 47 |
260840_at |
AT1G29050
|
TBL38, TRICHOME BIREFRINGENCE-LIKE 38 |
0.194 |
0.038 |
| 48 |
252470_at |
AT3G46930
|
[Protein kinase superfamily protein] |
0.194 |
0.496 |
| 49 |
251109_at |
AT5G01600
|
ATFER1, ARABIDOPSIS THALIANA FERRETIN 1, FER1, ferretin 1 |
0.193 |
0.217 |
| 50 |
254697_at |
AT4G17970
|
ALMT12, aluminum-activated, malate transporter 12, ATALMT12, QUAC1, quick-activating anion channel 1 |
0.189 |
-0.058 |
| 51 |
245740_at |
AT1G44100
|
AAP5, amino acid permease 5 |
0.189 |
-0.094 |
| 52 |
253172_at |
AT4G35060
|
HIPP25, heavy metal associated isoprenylated plant protein 25 |
0.189 |
0.444 |
| 53 |
249134_at |
AT5G43150
|
unknown |
0.188 |
-0.112 |
| 54 |
264879_at |
AT1G61260
|
unknown |
0.187 |
0.047 |
| 55 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.186 |
-0.039 |
| 56 |
249410_at |
AT5G40380
|
CRK42, cysteine-rich RLK (RECEPTOR-like protein kinase) 42 |
0.186 |
-0.166 |
| 57 |
264999_at |
AT1G67310
|
[Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains] |
0.185 |
-0.014 |
| 58 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.184 |
-0.586 |
| 59 |
251739_at |
AT3G56170
|
CAN, Ca-2+ dependent nuclease, CAN1, calcium dependent nuclease 1 |
0.183 |
0.275 |
| 60 |
255585_at |
AT4G01550
|
ANAC069, NAC domain containing protein 69, NAC069, NAC domain containing protein 69, NTM2, NAC with Transmembrane Motif 2 |
0.183 |
0.263 |
| 61 |
261832_at |
AT1G10650
|
[SBP (S-ribonuclease binding protein) family protein] |
0.182 |
-0.239 |
| 62 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.182 |
0.225 |
| 63 |
266480_at |
AT2G31130
|
unknown |
0.180 |
-0.001 |
| 64 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
0.177 |
0.081 |
| 65 |
251235_at |
AT3G62860
|
[alpha/beta-Hydrolases superfamily protein] |
0.176 |
0.205 |
| 66 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.173 |
0.336 |
| 67 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
0.172 |
0.227 |
| 68 |
257074_at |
AT3G19660
|
unknown |
0.171 |
0.569 |
| 69 |
258091_at |
AT3G14560
|
unknown |
0.170 |
0.432 |
| 70 |
261056_at |
AT1G01360
|
PYL9, PYRABACTIN RESISTANCE 1-LIKE 9, RCAR1, regulatory component of ABA receptor 1 |
0.169 |
-0.153 |
| 71 |
250811_at |
AT5G05110
|
[Cystatin/monellin family protein] |
0.169 |
0.192 |
| 72 |
249944_at |
AT5G22290
|
anac089, NAC domain containing protein 89, FSQ6, fructose-sensing quantitative trait locus 6, NAC089, NAC domain containing protein 89 |
0.169 |
0.136 |
| 73 |
259953_at |
AT1G74810
|
BOR5, REQUIRES HIGH BORON 5 |
0.165 |
-0.042 |
| 74 |
247577_at |
AT5G61290
|
[Flavin-binding monooxygenase family protein] |
0.164 |
-0.066 |
| 75 |
261399_at |
AT1G79620
|
[Leucine-rich repeat protein kinase family protein] |
0.163 |
-0.288 |
| 76 |
260054_at |
AT1G78130
|
UNE2, unfertilized embryo sac 2 |
0.163 |
0.157 |
| 77 |
250400_at |
AT5G10740
|
[Protein phosphatase 2C family protein] |
0.162 |
0.094 |
| 78 |
258119_at |
AT3G14720
|
ATMPK19, ARABIDOPSIS THALIANA MAP KINASE 19, MPK19, MAP kinase 19 |
0.159 |
-0.013 |
| 79 |
251011_at |
AT5G02560
|
HTA12, histone H2A 12 |
0.159 |
0.006 |
| 80 |
263805_at |
AT2G40400
|
unknown |
-0.451 |
-0.281 |
| 81 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
-0.384 |
0.221 |
| 82 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
-0.305 |
0.044 |
| 83 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.277 |
0.668 |
| 84 |
261684_at |
AT1G47400
|
unknown |
-0.273 |
-0.436 |
| 85 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
-0.243 |
0.136 |
| 86 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
-0.230 |
0.558 |
| 87 |
267462_at |
AT2G33735
|
[Chaperone DnaJ-domain superfamily protein] |
-0.224 |
-0.195 |
| 88 |
245411_at |
AT4G17240
|
unknown |
-0.224 |
0.299 |
| 89 |
264114_at |
AT2G31270
|
ATCDT1A, ARABIDOPSIS HOMOLOG OF YEAST CDT1 A, CDT1, ARABIDOPSIS HOMOLOG OF YEAST CDT1, CDT1A, homolog of yeast CDT1 A |
-0.220 |
-0.100 |
| 90 |
251538_at |
AT3G58660
|
[Ribosomal protein L1p/L10e family] |
-0.219 |
-0.072 |
| 91 |
246943_at |
AT5G25440
|
[Protein kinase superfamily protein] |
-0.217 |
0.419 |
| 92 |
264357_at |
AT1G03360
|
ATRRP4, ribosomal RNA processing 4, RRP4, ribosomal RNA processing 4 |
-0.208 |
-0.085 |
| 93 |
248065_at |
AT5G55580
|
[Mitochondrial transcription termination factor family protein] |
-0.207 |
-0.324 |
| 94 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-0.207 |
-0.524 |
| 95 |
250560_at |
AT5G08020
|
ATRPA70B, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT B, RPA70B, RPA70-kDa subunit B |
-0.204 |
-0.088 |
| 96 |
258828_at |
AT3G07130
|
ATPAP15, PURPLE ACID PHOSPHATASE 15, PAP15, purple acid phosphatase 15 |
-0.199 |
-0.129 |
| 97 |
258009_at |
AT3G19440
|
[Pseudouridine synthase family protein] |
-0.179 |
0.008 |
| 98 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
-0.173 |
0.640 |
| 99 |
254284_at |
AT4G22910
|
CCS52A1, cell cycle switch protein 52 A1, FZR2, FIZZY-related 2 |
-0.171 |
-0.086 |
| 100 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
-0.169 |
0.116 |
| 101 |
249637_at |
AT5G36900
|
unknown |
-0.167 |
-0.037 |
| 102 |
245154_at |
AT5G12460
|
unknown |
-0.166 |
-0.039 |
| 103 |
251271_at |
AT3G62050
|
[Putative endonuclease or glycosyl hydrolase] |
-0.166 |
-0.136 |
| 104 |
248295_at |
AT5G53070
|
[Ribosomal protein L9/RNase H1] |
-0.165 |
-0.073 |
| 105 |
263318_at |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
-0.164 |
-0.503 |
| 106 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
-0.162 |
0.503 |
| 107 |
259643_at |
AT1G68890
|
PHYLLO, PHYLLO |
-0.160 |
-0.147 |
| 108 |
253925_at |
AT4G26690
|
GDPDL3, Glycerophosphodiester phosphodiesterase (GDPD) like 3, GPDL2, GLYCEROPHOSPHODIESTERASE-LIKE 2, MRH5, MUTANT ROOT HAIR 5, SHV3, SHAVEN 3 |
-0.159 |
-0.033 |
| 109 |
248270_at |
AT5G53450
|
ORG1, OBP3-responsive gene 1 |
-0.157 |
-0.265 |
| 110 |
260667_at |
AT1G19440
|
KCS4, 3-ketoacyl-CoA synthase 4 |
-0.156 |
-0.145 |
| 111 |
250084_at |
AT5G17240
|
SDG40, SET domain group 40 |
-0.156 |
-0.112 |
| 112 |
256910_at |
AT3G24080
|
[KRR1 family protein] |
-0.155 |
-0.007 |
| 113 |
258898_at |
AT3G05740
|
RECQI1, RECQ helicase l1 |
-0.152 |
-0.077 |
| 114 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.152 |
-0.114 |
| 115 |
248841_at |
AT5G46740
|
UBP21, ubiquitin-specific protease 21 |
-0.151 |
-0.071 |
| 116 |
263435_at |
AT2G28600
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.150 |
0.018 |
| 117 |
252147_at |
AT3G51270
|
[protein serine/threonine kinases] |
-0.149 |
-0.074 |
| 118 |
253974_at |
AT4G26540
|
[Leucine-rich repeat receptor-like protein kinase family protein] |
-0.149 |
-0.206 |
| 119 |
245828_at |
AT1G57820
|
ORTH2, ORTHRUS 2, VIM1, VARIANT IN METHYLATION 1 |
-0.148 |
-0.052 |
| 120 |
245164_at |
AT2G33210
|
HSP60-2, heat shock protein 60-2 |
-0.147 |
-0.029 |
| 121 |
251593_at |
AT3G57660
|
NRPA1, nuclear RNA polymerase A1 |
-0.146 |
-0.176 |
| 122 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.146 |
-0.373 |
| 123 |
253973_at |
AT4G26555
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.145 |
-0.225 |
| 124 |
252684_at |
AT3G44400
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.144 |
0.183 |
| 125 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.143 |
-0.278 |
| 126 |
262696_at |
AT1G75870
|
unknown |
-0.143 |
-0.048 |
| 127 |
251378_at |
AT3G60660
|
unknown |
-0.142 |
-0.012 |
| 128 |
265250_at |
AT2G01950
|
BRL2, BRI1-like 2, VH1, VASCULAR HIGHWAY 1 |
-0.142 |
-0.409 |
| 129 |
260970_at |
AT1G53640
|
unknown |
-0.142 |
0.012 |
| 130 |
248776_at |
AT5G47900
|
unknown |
-0.142 |
-0.185 |
| 131 |
248691_at |
AT5G48310
|
unknown |
-0.141 |
-0.030 |
| 132 |
248528_at |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
-0.140 |
0.491 |
| 133 |
257621_at |
AT3G20410
|
CPK9, calmodulin-domain protein kinase 9 |
-0.139 |
0.371 |
| 134 |
250753_at |
AT5G05860
|
UGT76C2, UDP-glucosyl transferase 76C2 |
-0.138 |
0.036 |
| 135 |
263594_at |
AT2G01880
|
ATPAP7, PURPLE ACID PHOSPHATASE 7, PAP7, purple acid phosphatase 7 |
-0.138 |
0.048 |
| 136 |
246478_at |
AT5G15980
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.137 |
-0.113 |
| 137 |
257188_at |
AT3G13150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.136 |
0.003 |
| 138 |
251807_at |
AT3G55400
|
OVA1, OVULE ABORTION 1 |
-0.135 |
0.028 |
| 139 |
258871_at |
AT3G03060
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.133 |
-0.088 |
| 140 |
262657_at |
AT1G14210
|
[Ribonuclease T2 family protein] |
-0.132 |
-0.097 |
| 141 |
246094_at |
AT5G19300
|
unknown |
-0.131 |
-0.005 |
| 142 |
259703_at |
AT1G77790
|
[Glycosyl hydrolase superfamily protein] |
-0.131 |
-0.026 |
| 143 |
253592_at |
AT4G30840
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.130 |
-0.048 |
| 144 |
252574_at |
AT3G45430
|
LecRK-I.5, L-type lectin receptor kinase I.5 |
-0.129 |
-0.119 |
| 145 |
255282_at |
AT4G04600
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:CAA31653 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
-0.128 |
0.013 |
| 146 |
266957_at |
AT2G34640
|
HMR, HEMERA, PTAC12, plastid transcriptionally active 12, TAC12 |
-0.128 |
-0.114 |
| 147 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.127 |
-0.035 |
| 148 |
261032_at |
AT1G17430
|
[alpha/beta-Hydrolases superfamily protein] |
-0.126 |
0.006 |
| 149 |
266489_at |
AT2G35190
|
ATNPSN11, NPSN11, novel plant snare 11, NSPN11 |
-0.125 |
-0.134 |
| 150 |
245614_at |
AT4G14460
|
[copia-like retrotransposon family, has a 4.2e-253 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
-0.125 |
-0.018 |
| 151 |
251496_at |
AT3G59040
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.124 |
-0.203 |
| 152 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.124 |
0.220 |
| 153 |
256155_at |
AT3G08500
|
AtMYB83, myb domain protein 83, MYB83, myb domain protein 83 |
-0.124 |
-0.036 |
| 154 |
246559_at |
AT5G15550
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.123 |
-0.008 |
| 155 |
248456_at |
AT5G51380
|
[RNI-like superfamily protein] |
-0.123 |
-0.021 |
| 156 |
258013_at |
AT3G19320
|
[Leucine-rich repeat (LRR) family protein] |
-0.122 |
0.003 |
| 157 |
259378_at |
AT3G16310
|
1 |
-0.121 |
0.049 |