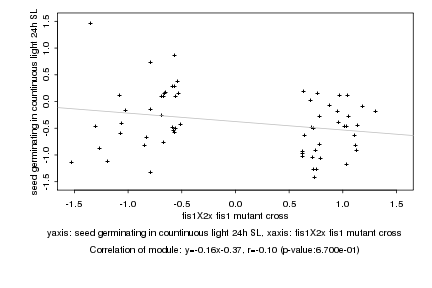
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
fis1X2x fis1 mutant cross |
Log2 signal ratio
seed germinating in countinuous light 24h SL |
| 1 |
254900_at |
AT4G11510
|
RALFL28, ralf-like 28 |
1.300 |
-0.185 |
| 2 |
250499_at |
AT5G09730
|
ATBX3, ATBXL3, BETA-XYLOSIDASE 3, BX3, BXL3, beta-xylosidase 3, XYL3 |
1.175 |
-0.081 |
| 3 |
252500_at |
AT3G46860
|
[Serine protease inhibitor, potato inhibitor I-type family protein] |
1.135 |
-0.432 |
| 4 |
267166_at |
AT2G37720
|
TBL15, TRICHOME BIREFRINGENCE-LIKE 15 |
1.123 |
-0.908 |
| 5 |
265517_at |
AT2G06090
|
[Plant self-incompatibility protein S1 family] |
1.116 |
-0.818 |
| 6 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.105 |
-0.630 |
| 7 |
247799_at |
AT5G58840
|
[Subtilase family protein] |
1.049 |
-0.280 |
| 8 |
257934_at |
AT3G25420
|
scpl21, serine carboxypeptidase-like 21 |
1.038 |
0.120 |
| 9 |
245738_at |
AT1G44130
|
[Eukaryotic aspartyl protease family protein] |
1.031 |
-0.462 |
| 10 |
253463_at |
AT4G32105
|
[Beta-1,3-N-Acetylglucosaminyltransferase family protein] |
1.027 |
-1.162 |
| 11 |
258130_at |
AT3G24510
|
[Defensin-like (DEFL) family protein] |
1.015 |
-0.458 |
| 12 |
266562_at |
AT2G23970
|
[Class I glutamine amidotransferase-like superfamily protein] |
0.961 |
0.130 |
| 13 |
259107_at |
AT3G05460
|
[sporozoite surface protein-related] |
0.952 |
-0.387 |
| 14 |
260066_at |
AT1G73610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.944 |
-0.186 |
| 15 |
250154_at |
AT5G15140
|
[Galactose mutarotase-like superfamily protein] |
0.871 |
-0.071 |
| 16 |
260396_at |
AT1G69720
|
HO3, heme oxygenase 3 |
0.784 |
-1.051 |
| 17 |
249500_at |
AT5G39260
|
ATEXP21, ATEXPA21, expansin A21, ATHEXP ALPHA 1.20, EXP21, EXPANSIN 21, EXPA21, expansin A21 |
0.776 |
-0.273 |
| 18 |
254801_at |
AT4G13080
|
XTH1, xyloglucan endotransglucosylase/hydrolase 1 |
0.775 |
-0.803 |
| 19 |
256600_at |
AT3G14850
|
TBL41, TRICHOME BIREFRINGENCE-LIKE 41 |
0.764 |
0.168 |
| 20 |
248202_at |
AT5G54220
|
[Cysteine-rich protein] |
0.749 |
-1.255 |
| 21 |
248275_at |
AT5G53520
|
ATOPT8, oligopeptide transporter 8, OPT8, oligopeptide transporter 8 |
0.737 |
-0.907 |
| 22 |
255956_at |
AT1G22015
|
DD46 |
0.728 |
-1.408 |
| 23 |
253532_at |
AT4G31570
|
unknown |
0.727 |
-0.486 |
| 24 |
254237_at |
AT4G23520
|
[Cysteine proteinases superfamily protein] |
0.722 |
-1.268 |
| 25 |
246834_at |
AT5G26630
|
[MADS-box transcription factor family protein] |
0.712 |
-1.033 |
| 26 |
256214_x_at |
AT1G51000
|
unknown |
0.700 |
-0.469 |
| 27 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
0.694 |
0.027 |
| 28 |
254566_at |
AT4G19240
|
unknown |
0.639 |
-0.624 |
| 29 |
254596_at |
AT4G18975
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.631 |
0.192 |
| 30 |
245754_at |
AT1G35183
|
unknown |
0.624 |
-0.931 |
| 31 |
256410_at |
AT1G66630
|
[Protein with RING/U-box and TRAF-like domains] |
0.619 |
-1.016 |
| 32 |
253655_at |
AT4G30070
|
LCR59, low-molecular-weight cysteine-rich 59 |
0.618 |
-0.955 |
| 33 |
249549_at |
AT5G38180
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.534 |
-1.129 |
| 34 |
255257_at |
AT4G05050
|
UBQ11, ubiquitin 11 |
-1.352 |
0.861 |
| 35 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-1.305 |
-0.455 |
| 36 |
258980_at |
AT3G08900
|
RGP, RGP3, reversibly glycosylated polypeptide 3 |
-1.267 |
-0.862 |
| 37 |
260326_at |
AT1G63910
|
AtMYB103, myb domain protein 103, MYB103, myb domain protein 103 |
-1.197 |
-1.111 |
| 38 |
263649_at |
AT1G04380
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.088 |
0.119 |
| 39 |
250611_at |
AT5G07200
|
ATGA20OX3, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 3, GA20OX3, gibberellin 20-oxidase 3, YAP169 |
-1.073 |
-0.582 |
| 40 |
259935_at |
AT1G71250
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.065 |
-0.400 |
| 41 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-1.028 |
-0.155 |
| 42 |
249479_at |
AT5G38960
|
[RmlC-like cupins superfamily protein] |
-0.855 |
-0.815 |
| 43 |
260265_at |
AT1G68510
|
LBD42, LOB domain-containing protein 42 |
-0.831 |
-0.659 |
| 44 |
256435_at |
AT3G11180
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.798 |
-1.320 |
| 45 |
259966_at |
AT1G76500
|
AHL29, AT-hook motif nuclear-localized protein 29, SOB3, SUPPRESSOR OF PHYB-4#3 |
-0.796 |
0.748 |
| 46 |
254452_at |
AT4G21100
|
DDB1B, damaged DNA binding protein 1B |
-0.792 |
-0.149 |
| 47 |
264377_at |
AT2G25060
|
AtENODL14, ENODL14, early nodulin-like protein 14 |
-0.690 |
0.112 |
| 48 |
263788_at |
AT2G24580
|
[FAD-dependent oxidoreductase family protein] |
-0.690 |
-0.253 |
| 49 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
-0.674 |
0.099 |
| 50 |
252078_at |
AT3G51740
|
IMK2, inflorescence meristem receptor-like kinase 2 |
-0.671 |
-0.756 |
| 51 |
244961_at |
ATCG01040
|
YCF5 |
-0.664 |
0.158 |
| 52 |
266917_at |
AT2G45830
|
DTA2, downstream target of AGL15 2 |
-0.657 |
0.184 |
| 53 |
267006_at |
AT2G34190
|
[Xanthine/uracil permease family protein] |
-0.593 |
0.299 |
| 54 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
-0.590 |
-0.474 |
| 55 |
258067_at |
AT3G25980
|
AtMAD2, MAD2, MITOTIC ARREST-DEFICIENT 2 |
-0.582 |
-0.525 |
| 56 |
247955_at |
AT5G56950
|
NAP1;3, nucleosome assembly protein 1;3, NFA03, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP A 03, NFA3, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP A3 |
-0.574 |
0.299 |
| 57 |
258480_at |
AT3G02640
|
unknown |
-0.570 |
0.865 |
| 58 |
254344_at |
AT4G22110
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.568 |
-0.576 |
| 59 |
259823_at |
AT1G66250
|
[O-Glycosyl hydrolases family 17 protein] |
-0.567 |
0.108 |
| 60 |
247425_at |
AT5G62550
|
unknown |
-0.559 |
-0.498 |
| 61 |
246505_at |
AT5G16250
|
unknown |
-0.549 |
0.378 |
| 62 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.537 |
0.156 |
| 63 |
253097_at |
AT4G37320
|
CYP81D5, cytochrome P450, family 81, subfamily D, polypeptide 5 |
-0.518 |
-0.412 |