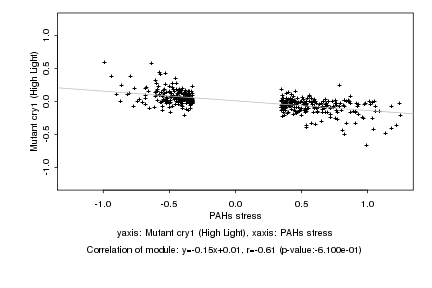
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
PAHs stress |
Log2 signal ratio
Mutant cry1 (High Light) |
| 1 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
1.246 |
-0.200 |
| 2 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.235 |
-0.018 |
| 3 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
1.218 |
-0.357 |
| 4 |
261684_at |
AT1G47400
|
unknown |
1.180 |
-0.065 |
| 5 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
1.175 |
-0.399 |
| 6 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
1.134 |
-0.471 |
| 7 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
1.083 |
-0.149 |
| 8 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
1.058 |
-0.141 |
| 9 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
1.057 |
-0.063 |
| 10 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
1.049 |
0.002 |
| 11 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
1.039 |
-0.410 |
| 12 |
248333_at |
AT5G52390
|
[PAR1 protein] |
1.037 |
-0.003 |
| 13 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
1.032 |
-0.257 |
| 14 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.020 |
-0.041 |
| 15 |
245692_at |
AT5G04150
|
BHLH101 |
1.011 |
0.001 |
| 16 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
0.987 |
-0.668 |
| 17 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
0.979 |
-0.030 |
| 18 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
0.969 |
-0.041 |
| 19 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.968 |
-0.250 |
| 20 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.961 |
-0.231 |
| 21 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.925 |
-0.040 |
| 22 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.924 |
-0.190 |
| 23 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.920 |
-0.067 |
| 24 |
248918_at |
AT5G45890
|
AtSAG12, SAG12, senescence-associated gene 12 |
0.909 |
-0.030 |
| 25 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
0.907 |
-0.159 |
| 26 |
264635_at |
AT1G65500
|
unknown |
0.907 |
-0.329 |
| 27 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
0.904 |
-0.139 |
| 28 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.889 |
-0.139 |
| 29 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.868 |
-0.020 |
| 30 |
253502_at |
AT4G31940
|
CYP82C4, cytochrome P450, family 82, subfamily C, polypeptide 4 |
0.868 |
0.079 |
| 31 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.865 |
-0.159 |
| 32 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.856 |
0.013 |
| 33 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
0.855 |
0.022 |
| 34 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.848 |
0.005 |
| 35 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.840 |
0.015 |
| 36 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.834 |
-0.322 |
| 37 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
0.824 |
-0.494 |
| 38 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
0.823 |
0.029 |
| 39 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.821 |
0.028 |
| 40 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.819 |
-0.071 |
| 41 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.815 |
-0.046 |
| 42 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.803 |
-0.427 |
| 43 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.800 |
-0.130 |
| 44 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
0.794 |
-0.027 |
| 45 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
0.781 |
0.257 |
| 46 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.780 |
-0.068 |
| 47 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.766 |
-0.159 |
| 48 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.766 |
-0.267 |
| 49 |
266486_at |
AT2G47950
|
unknown |
0.764 |
-0.017 |
| 50 |
255900_at |
AT1G17830
|
unknown |
0.762 |
-0.131 |
| 51 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.761 |
-0.019 |
| 52 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.758 |
-0.059 |
| 53 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
0.753 |
-0.252 |
| 54 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
0.751 |
0.021 |
| 55 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.751 |
-0.027 |
| 56 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.738 |
-0.074 |
| 57 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.738 |
-0.089 |
| 58 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.731 |
0.011 |
| 59 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.719 |
-0.230 |
| 60 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.715 |
-0.083 |
| 61 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.714 |
-0.096 |
| 62 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.712 |
-0.192 |
| 63 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
0.701 |
0.003 |
| 64 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.695 |
-0.046 |
| 65 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.691 |
-0.059 |
| 66 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.690 |
-0.164 |
| 67 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.686 |
-0.020 |
| 68 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.685 |
-0.075 |
| 69 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
0.679 |
0.037 |
| 70 |
260030_at |
AT1G68880
|
AtbZIP, basic leucine-zipper 8, bZIP, basic leucine-zipper 8 |
0.679 |
-0.153 |
| 71 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
0.676 |
-0.035 |
| 72 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.669 |
-0.098 |
| 73 |
249454_at |
AT5G39520
|
unknown |
0.662 |
0.006 |
| 74 |
266376_at |
AT2G14620
|
XTH10, xyloglucan endotransglucosylase/hydrolase 10 |
0.659 |
0.012 |
| 75 |
252958_at |
AT4G38620
|
ATMYB4, myb domain protein 4, MYB4, myb domain protein 4 |
0.658 |
-0.061 |
| 76 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.658 |
-0.111 |
| 77 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.655 |
-0.083 |
| 78 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.652 |
-0.165 |
| 79 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.651 |
-0.303 |
| 80 |
250702_at |
AT5G06730
|
[Peroxidase superfamily protein] |
0.644 |
-0.015 |
| 81 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.629 |
-0.043 |
| 82 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.627 |
-0.081 |
| 83 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
0.617 |
-0.162 |
| 84 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.615 |
-0.140 |
| 85 |
252888_at |
AT4G39210
|
APL3 |
0.609 |
-0.106 |
| 86 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.607 |
-0.018 |
| 87 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
0.603 |
0.004 |
| 88 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.599 |
-0.339 |
| 89 |
249476_at |
AT5G38910
|
[RmlC-like cupins superfamily protein] |
0.598 |
0.001 |
| 90 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.598 |
-0.066 |
| 91 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.596 |
-0.078 |
| 92 |
267391_at |
AT2G44480
|
BGLU17, beta glucosidase 17 |
0.596 |
-0.071 |
| 93 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
0.594 |
-0.098 |
| 94 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.591 |
0.045 |
| 95 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
0.587 |
-0.161 |
| 96 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
0.585 |
0.015 |
| 97 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.581 |
-0.034 |
| 98 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.577 |
-0.010 |
| 99 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.572 |
-0.319 |
| 100 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.570 |
0.048 |
| 101 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.568 |
0.000 |
| 102 |
256603_at |
AT3G28270
|
unknown |
0.560 |
0.106 |
| 103 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
0.559 |
0.003 |
| 104 |
251545_at |
AT3G58810
|
ATMTP3, ARABIDOPSIS METAL TOLERANCE PROTEIN 3, ATMTPA2, MTP3, METAL TOLERANCE PROTEIN 3, MTPA2, metal tolerance protein A2 |
0.558 |
0.084 |
| 105 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.552 |
-0.103 |
| 106 |
248700_at |
AT5G48400
|
ATGLR1.2, GLR1.2, GLUTAMATE RECEPTOR 1.2 |
0.542 |
0.059 |
| 107 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.541 |
-0.072 |
| 108 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.539 |
-0.101 |
| 109 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
0.537 |
-0.044 |
| 110 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.536 |
-0.141 |
| 111 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
0.534 |
-0.023 |
| 112 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.533 |
-0.393 |
| 113 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
0.532 |
-0.018 |
| 114 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.532 |
-0.352 |
| 115 |
264912_at |
AT1G60750
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.531 |
0.002 |
| 116 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.530 |
-0.117 |
| 117 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.528 |
-0.155 |
| 118 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.528 |
-0.099 |
| 119 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.524 |
-0.060 |
| 120 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.524 |
-0.136 |
| 121 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
0.517 |
-0.122 |
| 122 |
264375_at |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
0.516 |
-0.007 |
| 123 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.515 |
-0.026 |
| 124 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
0.512 |
-0.032 |
| 125 |
249495_at |
AT5G39100
|
GLP6, germin-like protein 6 |
0.508 |
0.039 |
| 126 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
0.503 |
0.071 |
| 127 |
266269_at |
AT2G29480
|
ATGSTU2, glutathione S-transferase tau 2, GST20, GLUTATHIONE S-TRANSFERASE 20, GSTU2, glutathione S-transferase tau 2 |
0.497 |
0.004 |
| 128 |
260492_at |
AT2G41850
|
ADPG2, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2, PGAZAT, polygalacturonase abscission zone A. thaliana |
0.494 |
-0.092 |
| 129 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
0.494 |
-0.203 |
| 130 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
0.489 |
-0.130 |
| 131 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
0.487 |
-0.005 |
| 132 |
251293_at |
AT3G61930
|
unknown |
0.485 |
-0.002 |
| 133 |
264958_at |
AT1G76960
|
unknown |
0.483 |
-0.129 |
| 134 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.480 |
-0.148 |
| 135 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
0.474 |
-0.002 |
| 136 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.471 |
-0.085 |
| 137 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.468 |
-0.016 |
| 138 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.466 |
-0.159 |
| 139 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
0.465 |
-0.019 |
| 140 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
0.463 |
0.055 |
| 141 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.460 |
-0.035 |
| 142 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.460 |
-0.177 |
| 143 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.455 |
-0.088 |
| 144 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
0.453 |
0.163 |
| 145 |
260933_at |
AT1G02470
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.449 |
0.004 |
| 146 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.448 |
-0.139 |
| 147 |
246145_at |
AT5G19880
|
[Peroxidase superfamily protein] |
0.446 |
0.023 |
| 148 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
0.445 |
-0.001 |
| 149 |
264717_at |
AT1G70140
|
ATFH8, formin 8, FH8, formin 8 |
0.444 |
-0.077 |
| 150 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
0.441 |
0.013 |
| 151 |
249705_at |
AT5G35580
|
[Protein kinase superfamily protein] |
0.441 |
-0.044 |
| 152 |
247602_at |
AT5G60900
|
RLK1, receptor-like protein kinase 1 |
0.440 |
-0.128 |
| 153 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.434 |
0.079 |
| 154 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.434 |
-0.195 |
| 155 |
260517_at |
AT1G51420
|
ATSPP1, SUCROSE-PHOSPHATASE 1, SPP1, sucrose-phosphatase 1 |
0.430 |
0.033 |
| 156 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.429 |
-0.060 |
| 157 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
0.428 |
0.026 |
| 158 |
260563_at |
AT2G43840
|
UGT74F1, UDP-glycosyltransferase 74 F1 |
0.427 |
-0.013 |
| 159 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.426 |
0.045 |
| 160 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.423 |
-0.051 |
| 161 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.420 |
-0.048 |
| 162 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.420 |
-0.112 |
| 163 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
0.420 |
0.119 |
| 164 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
0.420 |
-0.010 |
| 165 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.420 |
-0.010 |
| 166 |
252572_at |
AT3G45290
|
ATMLO3, MILDEW RESISTANCE LOCUS O 3, MLO3, MILDEW RESISTANCE LOCUS O 3 |
0.418 |
0.018 |
| 167 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
0.416 |
-0.012 |
| 168 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
0.415 |
-0.074 |
| 169 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.415 |
-0.006 |
| 170 |
264751_at |
AT1G23020
|
ATFRO3, FERRIC REDUCTION OXIDASE 3, FRO3, ferric reduction oxidase 3 |
0.414 |
-0.026 |
| 171 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.413 |
0.054 |
| 172 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.412 |
-0.058 |
| 173 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.411 |
-0.057 |
| 174 |
266851_at |
AT2G26820
|
ATPP2-A3, phloem protein 2-A3, PP2-A3, phloem protein 2-A3 |
0.407 |
-0.004 |
| 175 |
267345_at |
AT2G44240
|
unknown |
0.405 |
-0.057 |
| 176 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.402 |
-0.067 |
| 177 |
258209_at |
AT3G14060
|
unknown |
0.400 |
-0.161 |
| 178 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
0.400 |
0.146 |
| 179 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
0.398 |
0.039 |
| 180 |
257348_at |
AT2G42140
|
[VQ motif-containing protein] |
0.393 |
-0.042 |
| 181 |
252383_at |
AT3G47780
|
ABCA7, ATP-binding cassette A7, ATATH6, A. THALIANA ABC2 HOMOLOG 6, ATH6, ABC2 homolog 6 |
0.392 |
-0.176 |
| 182 |
254416_at |
AT4G21380
|
ARK3, receptor kinase 3, RK3, receptor kinase 3 |
0.390 |
-0.039 |
| 183 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.389 |
-0.036 |
| 184 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
0.385 |
0.136 |
| 185 |
248979_at |
AT5G45080
|
AtPP2-A6, phloem protein 2-A6, PP2-A6, phloem protein 2-A6 |
0.382 |
0.009 |
| 186 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.382 |
0.055 |
| 187 |
256366_at |
AT1G66880
|
[Protein kinase superfamily protein] |
0.381 |
-0.080 |
| 188 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.381 |
0.023 |
| 189 |
267300_at |
AT2G30140
|
UGT87A2, UDP-glucosyl transferase 87A2 |
0.378 |
-0.052 |
| 190 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.377 |
-0.132 |
| 191 |
259251_at |
AT3G07600
|
[Heavy metal transport/detoxification superfamily protein ] |
0.376 |
0.010 |
| 192 |
248889_at |
AT5G46230
|
unknown |
0.374 |
-0.041 |
| 193 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.373 |
-0.030 |
| 194 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
0.373 |
-0.096 |
| 195 |
260005_at |
AT1G67920
|
unknown |
0.369 |
-0.023 |
| 196 |
260522_x_at |
AT2G41730
|
unknown |
0.367 |
-0.202 |
| 197 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
0.366 |
-0.080 |
| 198 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.364 |
-0.004 |
| 199 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
0.362 |
-0.119 |
| 200 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.362 |
0.060 |
| 201 |
258203_at |
AT3G13950
|
unknown |
0.362 |
-0.017 |
| 202 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.357 |
-0.161 |
| 203 |
267288_at |
AT2G23680
|
[Cold acclimation protein WCOR413 family] |
0.356 |
-0.063 |
| 204 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.355 |
-0.041 |
| 205 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
0.355 |
-0.109 |
| 206 |
254318_at |
AT4G22530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.354 |
0.030 |
| 207 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
0.354 |
0.028 |
| 208 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.352 |
-0.032 |
| 209 |
253779_at |
AT4G28490
|
HAE, HAESA, RLK5, RECEPTOR-LIKE PROTEIN KINASE 5 |
0.351 |
0.099 |
| 210 |
251858_at |
AT3G54820
|
PIP2;5, plasma membrane intrinsic protein 2;5, PIP2D, PLASMA MEMBRANE INTRINSIC PROTEIN 2D |
0.350 |
-0.050 |
| 211 |
267550_at |
AT2G32800
|
AP4.3A, LecRK-S.2, L-type lectin receptor kinase S.2 |
0.350 |
0.051 |
| 212 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.349 |
-0.216 |
| 213 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.348 |
-0.078 |
| 214 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.348 |
-0.090 |
| 215 |
257846_at |
AT3G12910
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
0.347 |
0.020 |
| 216 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
0.346 |
-0.058 |
| 217 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.345 |
-0.124 |
| 218 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
0.344 |
0.184 |
| 219 |
263565_at |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
0.344 |
0.003 |
| 220 |
264929_at |
AT1G60730
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.343 |
-0.078 |
| 221 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.991 |
0.596 |
| 222 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.943 |
0.384 |
| 223 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.901 |
0.116 |
| 224 |
248028_at |
AT5G55620
|
unknown |
-0.876 |
0.009 |
| 225 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.865 |
0.244 |
| 226 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
-0.821 |
0.109 |
| 227 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.808 |
0.131 |
| 228 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.800 |
0.385 |
| 229 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.778 |
-0.061 |
| 230 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.767 |
0.205 |
| 231 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.742 |
0.002 |
| 232 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.732 |
0.037 |
| 233 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.707 |
-0.007 |
| 234 |
250936_at |
AT5G03120
|
unknown |
-0.688 |
0.095 |
| 235 |
246860_at |
AT5G25840
|
unknown |
-0.687 |
0.057 |
| 236 |
261480_at |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-0.687 |
0.208 |
| 237 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
-0.685 |
-0.032 |
| 238 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.676 |
0.217 |
| 239 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-0.665 |
0.162 |
| 240 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.653 |
-0.100 |
| 241 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.641 |
0.578 |
| 242 |
258897_at |
AT3G05730
|
[LOCATED IN: endomembrane system] |
-0.615 |
-0.078 |
| 243 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.612 |
0.323 |
| 244 |
250189_at |
AT5G14410
|
unknown |
-0.610 |
0.082 |
| 245 |
254221_at |
AT4G23820
|
[Pectin lyase-like superfamily protein] |
-0.609 |
0.150 |
| 246 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.604 |
0.095 |
| 247 |
267158_at |
AT2G37640
|
ATEXP3, EXPANSIN 3, ATEXPA3, ARABIDOPSIS THALIANA EXPANSIN A3, ATHEXP ALPHA 1.9, EXP3 |
-0.602 |
0.156 |
| 248 |
252950_at |
AT4G38690
|
[PLC-like phosphodiesterases superfamily protein] |
-0.602 |
0.278 |
| 249 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.596 |
0.161 |
| 250 |
256096_at |
AT1G13650
|
unknown |
-0.593 |
0.197 |
| 251 |
253372_at |
AT4G33220
|
ATPME44, A. THALIANA PECTIN METHYLESTERASE 44, PME44, pectin methylesterase 44 |
-0.591 |
0.023 |
| 252 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.585 |
0.237 |
| 253 |
267262_at |
AT2G22990
|
SCPL8, SERINE CARBOXYPEPTIDASE-LIKE 8, SNG1, sinapoylglucose 1 |
-0.582 |
0.111 |
| 254 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.580 |
0.444 |
| 255 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.577 |
0.096 |
| 256 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.572 |
-0.008 |
| 257 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
-0.570 |
0.417 |
| 258 |
267626_at |
AT2G42250
|
CYP712A1, cytochrome P450, family 712, subfamily A, polypeptide 1 |
-0.567 |
-0.003 |
| 259 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.562 |
0.150 |
| 260 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.559 |
0.010 |
| 261 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.558 |
0.082 |
| 262 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
-0.554 |
-0.075 |
| 263 |
262211_at |
AT1G74930
|
ORA47 |
-0.553 |
-0.132 |
| 264 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.552 |
0.004 |
| 265 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.551 |
0.175 |
| 266 |
263674_at |
AT2G04790
|
unknown |
-0.546 |
0.044 |
| 267 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.546 |
0.092 |
| 268 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.540 |
0.137 |
| 269 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.539 |
0.193 |
| 270 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
-0.538 |
0.025 |
| 271 |
260150_at |
AT1G52820
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.535 |
0.017 |
| 272 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
-0.535 |
0.011 |
| 273 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.533 |
0.427 |
| 274 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.531 |
0.005 |
| 275 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
-0.530 |
0.266 |
| 276 |
254098_at |
AT4G25100
|
ATFSD1, ARABIDOPSIS FE SUPEROXIDE DISMUTASE 1, FSD1, Fe superoxide dismutase 1 |
-0.529 |
-0.019 |
| 277 |
245088_at |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
-0.528 |
0.072 |
| 278 |
260038_at |
AT1G68875
|
unknown |
-0.526 |
0.018 |
| 279 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.526 |
0.145 |
| 280 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
-0.518 |
0.010 |
| 281 |
254638_at |
AT4G18740
|
[Rho termination factor] |
-0.515 |
0.049 |
| 282 |
257925_at |
AT3G23170
|
unknown |
-0.511 |
-0.013 |
| 283 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.506 |
0.148 |
| 284 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-0.503 |
-0.089 |
| 285 |
260012_at |
AT1G67865
|
unknown |
-0.501 |
0.167 |
| 286 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
-0.501 |
-0.030 |
| 287 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.499 |
0.231 |
| 288 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.497 |
-0.004 |
| 289 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.495 |
-0.157 |
| 290 |
250257_at |
AT5G13770
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.493 |
0.026 |
| 291 |
258708_at |
AT3G09580
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.492 |
0.080 |
| 292 |
253425_at |
AT4G32190
|
[Myosin heavy chain-related protein] |
-0.490 |
0.074 |
| 293 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.490 |
0.122 |
| 294 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.488 |
0.086 |
| 295 |
246792_at |
AT5G27290
|
unknown |
-0.484 |
0.062 |
| 296 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.483 |
0.228 |
| 297 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.483 |
-0.072 |
| 298 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-0.482 |
0.278 |
| 299 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.473 |
0.103 |
| 300 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.473 |
0.191 |
| 301 |
267509_at |
AT2G45660
|
AGL20, AGAMOUS-like 20, ATSOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1, SOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1 |
-0.468 |
0.153 |
| 302 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.463 |
0.032 |
| 303 |
248509_at |
AT5G50335
|
unknown |
-0.460 |
0.356 |
| 304 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.459 |
0.064 |
| 305 |
263142_at |
AT1G65230
|
[Uncharacterized conserved protein (DUF2358)] |
-0.458 |
0.055 |
| 306 |
261141_at |
AT1G19740
|
[ATP-dependent protease La (LON) domain protein] |
-0.457 |
0.047 |
| 307 |
251353_at |
AT3G61080
|
[Protein kinase superfamily protein] |
-0.455 |
0.059 |
| 308 |
265184_at |
AT1G23710
|
unknown |
-0.455 |
-0.039 |
| 309 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
-0.453 |
0.079 |
| 310 |
265400_at |
AT2G10940
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.453 |
0.077 |
| 311 |
253495_at |
AT4G31850
|
PGR3, proton gradient regulation 3 |
-0.452 |
0.014 |
| 312 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.452 |
0.279 |
| 313 |
245444_at |
AT4G16740
|
ATTPS03, terpene synthase 03, TPS03, terpene synthase 03 |
-0.452 |
0.081 |
| 314 |
245318_at |
AT4G16980
|
[arabinogalactan-protein family] |
-0.449 |
0.045 |
| 315 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.447 |
0.186 |
| 316 |
251142_at |
AT5G01015
|
unknown |
-0.443 |
0.131 |
| 317 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.440 |
0.084 |
| 318 |
247347_at |
AT5G63780
|
SHA1, shoot apical meristem arrest 1 |
-0.436 |
0.039 |
| 319 |
250167_at |
AT5G15310
|
ATMIXTA, ATMYB16, myb domain protein 16, MYB16, myb domain protein 16 |
-0.436 |
0.137 |
| 320 |
246487_at |
AT5G16030
|
unknown |
-0.434 |
0.168 |
| 321 |
250073_at |
AT5G17170
|
ENH1, enhancer of sos3-1 |
-0.434 |
-0.011 |
| 322 |
258021_at |
AT3G19380
|
PUB25, plant U-box 25 |
-0.434 |
0.011 |
| 323 |
259658_at |
AT1G55370
|
NDF5, NDH-dependent cyclic electron flow 5 |
-0.432 |
-0.041 |
| 324 |
252280_at |
AT3G49260
|
iqd21, IQ-domain 21 |
-0.432 |
0.065 |
| 325 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.431 |
0.040 |
| 326 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-0.430 |
0.090 |
| 327 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
-0.430 |
0.188 |
| 328 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
-0.430 |
0.041 |
| 329 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.426 |
0.141 |
| 330 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-0.425 |
0.017 |
| 331 |
259791_at |
AT1G29700
|
[Metallo-hydrolase/oxidoreductase superfamily protein] |
-0.424 |
0.009 |
| 332 |
262970_at |
AT1G75690
|
LQY1, LOW QUANTUM YIELD OF PHOTOSYSTEM II 1 |
-0.421 |
0.035 |
| 333 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
-0.419 |
0.070 |
| 334 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-0.417 |
-0.005 |
| 335 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.417 |
0.035 |
| 336 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.416 |
0.167 |
| 337 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.416 |
-0.006 |
| 338 |
251906_at |
AT3G53720
|
ATCHX20, cation/H+ exchanger 20, CHX20, cation/H+ exchanger 20 |
-0.413 |
-0.054 |
| 339 |
250031_at |
AT5G18240
|
ATMYR1, ARABIDOPSIS MYB-RELATED PROTEIN 1, MYR1, myb-related protein 1 |
-0.412 |
0.011 |
| 340 |
265248_at |
AT2G43010
|
AtPIF4, PIF4, phytochrome interacting factor 4, SRL2 |
-0.412 |
0.168 |
| 341 |
265265_at |
AT2G42900
|
[Plant basic secretory protein (BSP) family protein] |
-0.411 |
0.133 |
| 342 |
252853_at |
AT4G39710
|
FKBP16-2, FK506-binding protein 16-2, PnsL4, Photosynthetic NDH subcomplex L 4 |
-0.410 |
0.057 |
| 343 |
246019_at |
AT5G10690
|
[pentatricopeptide (PPR) repeat-containing protein / CBS domain-containing protein] |
-0.406 |
-0.027 |
| 344 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.406 |
0.052 |
| 345 |
266293_at |
AT2G29360
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.403 |
-0.067 |
| 346 |
246200_at |
AT4G37240
|
unknown |
-0.402 |
-0.091 |
| 347 |
265454_at |
AT2G46530
|
ARF11, auxin response factor 11 |
-0.402 |
0.199 |
| 348 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
-0.399 |
0.117 |
| 349 |
253412_at |
AT4G33000
|
ATCBL10, CBL10, calcineurin B-like protein 10, SCABP8, SOS3-LIKE CALCIUM BINDING PROTEIN 8 |
-0.399 |
0.036 |
| 350 |
262598_at |
AT1G15260
|
unknown |
-0.398 |
0.068 |
| 351 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
-0.394 |
0.023 |
| 352 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.392 |
0.149 |
| 353 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
-0.390 |
0.028 |
| 354 |
264872_at |
AT1G24260
|
AGL9, AGAMOUS-like 9, SEP3, SEPALLATA3 |
-0.387 |
-0.204 |
| 355 |
247486_at |
AT5G62140
|
unknown |
-0.387 |
0.027 |
| 356 |
266805_at |
AT2G30010
|
TBL45, TRICHOME BIREFRINGENCE-LIKE 45 |
-0.386 |
0.071 |
| 357 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-0.385 |
0.072 |
| 358 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.382 |
0.036 |
| 359 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-0.382 |
0.038 |
| 360 |
260877_at |
AT1G21500
|
unknown |
-0.381 |
0.089 |
| 361 |
266278_at |
AT2G29300
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.381 |
0.174 |
| 362 |
263915_at |
AT2G36430
|
unknown |
-0.378 |
0.061 |
| 363 |
245730_at |
AT1G73470
|
unknown |
-0.378 |
0.046 |
| 364 |
259163_at |
AT3G01490
|
[Protein kinase superfamily protein] |
-0.377 |
0.119 |
| 365 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
-0.377 |
-0.114 |
| 366 |
251024_at |
AT5G02180
|
[Transmembrane amino acid transporter family protein] |
-0.376 |
-0.022 |
| 367 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.376 |
0.096 |
| 368 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.376 |
0.134 |
| 369 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.375 |
0.029 |
| 370 |
251068_at |
AT5G01920
|
STN8, State transition 8 |
-0.373 |
0.034 |
| 371 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.371 |
0.100 |
| 372 |
252118_at |
AT3G51400
|
unknown |
-0.370 |
0.009 |
| 373 |
245010_at |
ATCG00420
|
NDHJ, NADH dehydrogenase subunit J |
-0.369 |
0.052 |
| 374 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
-0.368 |
0.043 |
| 375 |
261118_at |
AT1G75460
|
[ATP-dependent protease La (LON) domain protein] |
-0.364 |
0.109 |
| 376 |
252463_at |
AT3G47070
|
[LOCATED IN: thylakoid, chloroplast thylakoid membrane, chloroplast, chloroplast envelope] |
-0.363 |
0.005 |
| 377 |
253440_at |
AT4G32570
|
TIFY8, TIFY domain protein 8 |
-0.361 |
0.087 |
| 378 |
256130_at |
AT1G18170
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.360 |
0.039 |
| 379 |
260769_at |
AT1G49010
|
[Duplicated homeodomain-like superfamily protein] |
-0.359 |
-0.004 |
| 380 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.359 |
0.083 |
| 381 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
-0.359 |
-0.135 |
| 382 |
262151_at |
AT1G52510
|
[alpha/beta-Hydrolases superfamily protein] |
-0.358 |
-0.014 |
| 383 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-0.358 |
0.164 |
| 384 |
265877_at |
AT2G42380
|
ATBZIP34, BZIP34 |
-0.357 |
0.109 |
| 385 |
251036_at |
AT5G02160
|
unknown |
-0.357 |
0.063 |
| 386 |
258156_at |
AT3G18050
|
unknown |
-0.356 |
0.091 |
| 387 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.354 |
0.055 |
| 388 |
259207_at |
AT3G09050
|
unknown |
-0.354 |
-0.022 |
| 389 |
248624_at |
AT5G48790
|
unknown |
-0.353 |
0.050 |
| 390 |
264096_at |
AT1G78995
|
unknown |
-0.353 |
0.016 |
| 391 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.353 |
0.094 |
| 392 |
264201_at |
AT1G22630
|
unknown |
-0.353 |
0.002 |
| 393 |
262569_at |
AT1G15180
|
[MATE efflux family protein] |
-0.353 |
0.066 |
| 394 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
-0.352 |
0.122 |
| 395 |
247977_at |
AT5G56850
|
unknown |
-0.352 |
-0.013 |
| 396 |
257954_at |
AT3G21760
|
HYR1, HYPOSTATIN RESISTANCE 1 |
-0.351 |
0.006 |
| 397 |
247921_at |
AT5G57660
|
ATCOL5, BBX6, B-box domain protein 6, COL5, CONSTANS-like 5 |
-0.351 |
0.114 |
| 398 |
246159_at |
AT5G20935
|
CRR42, CHLORORESPIRATORY REDUCTION 42 |
-0.351 |
-0.024 |
| 399 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
-0.351 |
0.051 |
| 400 |
252876_at |
AT4G39970
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.350 |
0.002 |
| 401 |
245383_at |
AT4G17810
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.349 |
0.128 |
| 402 |
258495_at |
AT3G02690
|
[nodulin MtN21 /EamA-like transporter family protein] |
-0.348 |
0.019 |
| 403 |
259140_at |
AT3G10230
|
AtLCY, LYC, lycopene cyclase |
-0.348 |
0.026 |
| 404 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.348 |
0.105 |
| 405 |
250857_at |
AT5G04790
|
unknown |
-0.348 |
0.157 |
| 406 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
-0.347 |
-0.096 |
| 407 |
252412_at |
AT3G47295
|
unknown |
-0.345 |
0.024 |
| 408 |
248491_at |
AT5G51010
|
[Rubredoxin-like superfamily protein] |
-0.344 |
-0.002 |
| 409 |
250075_at |
AT5G17670
|
[alpha/beta-Hydrolases superfamily protein] |
-0.344 |
-0.025 |
| 410 |
256613_at |
AT3G29290
|
emb2076, embryo defective 2076 |
-0.343 |
-0.018 |
| 411 |
263597_at |
AT2G01870
|
unknown |
-0.343 |
-0.016 |
| 412 |
248870_at |
AT5G46710
|
[PLATZ transcription factor family protein] |
-0.343 |
-0.060 |
| 413 |
250243_at |
AT5G13630
|
ABAR, ABA-BINDING PROTEIN, CCH, CONDITIONAL CHLORINA, CCH1, CHLH, H SUBUNIT OF MG-CHELATASE, GUN5, GENOMES UNCOUPLED 5 |
-0.342 |
-0.003 |
| 414 |
256655_at |
AT3G18890
|
AtTic62, translocon at the inner envelope membrane of chloroplasts 62, Tic62, translocon at the inner envelope membrane of chloroplasts 62 |
-0.341 |
0.014 |
| 415 |
260968_at |
AT1G12250
|
[Pentapeptide repeat-containing protein] |
-0.340 |
0.025 |
| 416 |
248329_at |
AT5G52780
|
unknown |
-0.339 |
0.021 |
| 417 |
267057_at |
AT2G32500
|
[Stress responsive alpha-beta barrel domain protein] |
-0.338 |
-0.029 |
| 418 |
259460_at |
AT1G44000
|
unknown |
-0.338 |
0.022 |
| 419 |
258025_at |
AT3G19480
|
3-PGDH, PGDH3, phosphoglycerate dehydrogenase 3 |
-0.337 |
0.015 |
| 420 |
257772_at |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.337 |
0.160 |
| 421 |
249510_at |
AT5G38510
|
[Rhomboid-related intramembrane serine protease family protein] |
-0.337 |
0.005 |
| 422 |
256514_at |
AT1G66130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.336 |
0.057 |
| 423 |
246510_at |
AT5G15410
|
ATCNGC2, CYCLIC NUCLEOTIDE-GATED CHANNEL 2, CNGC2, CYCLIC NUCLEOTIDE GATED CHANNEL 2, DND1, DEFENSE NO DEATH 1 |
-0.336 |
0.036 |
| 424 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.335 |
0.029 |
| 425 |
258392_at |
AT3G15400
|
ATA20, anther 20 |
-0.334 |
0.054 |
| 426 |
262612_at |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.333 |
0.002 |
| 427 |
253283_at |
AT4G34090
|
unknown |
-0.332 |
-0.006 |
| 428 |
251931_at |
AT3G53950
|
[glyoxal oxidase-related protein] |
-0.332 |
0.036 |
| 429 |
262175_at |
AT1G74880
|
NDH-O, NAD(P)H:plastoquinone dehydrogenase complex subunit O, NdhO, NADH dehydrogenase-like complex ) |
-0.332 |
0.011 |
| 430 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.332 |
0.240 |
| 431 |
261941_at |
AT1G22490
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.331 |
-0.019 |
| 432 |
266572_at |
AT2G23840
|
[HNH endonuclease] |
-0.331 |
0.021 |
| 433 |
246531_at |
AT5G15800
|
AGL2, AGAMOUS-like 2, SEP1, SEPALLATA1 |
-0.331 |
0.051 |
| 434 |
247734_at |
AT5G59400
|
unknown |
-0.330 |
-0.024 |
| 435 |
260041_at |
AT1G68780
|
[RNI-like superfamily protein] |
-0.329 |
0.128 |
| 436 |
252092_at |
AT3G51420
|
ATSSL4, STRICTOSIDINE SYNTHASE-LIKE 4, SSL4, strictosidine synthase-like 4 |
-0.328 |
0.022 |
| 437 |
250133_at |
AT5G16400
|
ATF2, TRXF2, thioredoxin F2 |
-0.327 |
0.094 |
| 438 |
265742_at |
AT2G01290
|
RPI2, ribose-5-phosphate isomerase 2 |
-0.326 |
0.005 |
| 439 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.325 |
-0.011 |