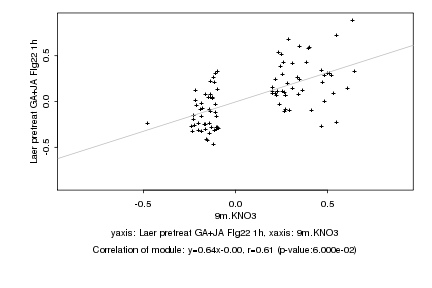
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
9m.KNO3 |
Log2 signal ratio
Laer pretreat GA+JA Flg22 1h |
| 1 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.646 |
0.330 |
| 2 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
0.636 |
0.895 |
| 3 |
246250_at |
AT4G36880
|
CP1, cysteine proteinase1, RDL1, RD21A-LIKE PROTEASE1 |
0.605 |
0.148 |
| 4 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
0.545 |
-0.221 |
| 5 |
261475_at |
AT1G14550
|
[Peroxidase superfamily protein] |
0.544 |
0.723 |
| 6 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
0.530 |
0.088 |
| 7 |
260394_at |
AT1G74080
|
ATMYB122, MYB DOMAIN PROTEIN 122, MYB122, myb domain protein 122 |
0.518 |
0.288 |
| 8 |
246145_at |
AT5G19880
|
[Peroxidase superfamily protein] |
0.509 |
0.312 |
| 9 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.496 |
0.308 |
| 10 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.484 |
0.011 |
| 11 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.482 |
0.287 |
| 12 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.473 |
0.210 |
| 13 |
250472_at |
AT5G10210
|
unknown |
0.467 |
-0.263 |
| 14 |
255945_at |
AT5G28610
|
unknown |
0.463 |
0.339 |
| 15 |
246996_at |
AT5G67420
|
ASL39, ASYMMETRIC LEAVES2-LIKE 39, LBD37, LOB domain-containing protein 37 |
0.410 |
-0.094 |
| 16 |
248316_at |
AT5G52670
|
[Copper transport protein family] |
0.398 |
0.600 |
| 17 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
0.395 |
0.587 |
| 18 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.386 |
0.426 |
| 19 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
0.360 |
0.126 |
| 20 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.345 |
0.242 |
| 21 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
0.344 |
0.611 |
| 22 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.338 |
0.082 |
| 23 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.335 |
0.265 |
| 24 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.309 |
0.422 |
| 25 |
255734_at |
AT1G25550
|
[myb-like transcription factor family protein] |
0.306 |
0.147 |
| 26 |
259961_at |
AT1G53700
|
PK3AT, PROTEIN KINASE 3 ARABIDOPSIS THALIANA, WAG1, WAG 1 |
0.293 |
-0.094 |
| 27 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.283 |
0.682 |
| 28 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.278 |
0.200 |
| 29 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.271 |
0.076 |
| 30 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.269 |
-0.081 |
| 31 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.267 |
0.075 |
| 32 |
262981_at |
AT1G75590
|
SAUR52, SMALL AUXIN UPREGULATED RNA 52 |
0.262 |
0.102 |
| 33 |
254027_at |
AT4G25835
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.262 |
-0.100 |
| 34 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
0.261 |
0.433 |
| 35 |
262671_at |
AT1G76040
|
CPK29, calcium-dependent protein kinase 29 |
0.254 |
0.303 |
| 36 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
0.252 |
0.110 |
| 37 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.250 |
0.516 |
| 38 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
0.240 |
0.389 |
| 39 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.235 |
-0.031 |
| 40 |
247145_at |
AT5G65600
|
LecRK-IX.2, L-type lectin receptor kinase IX.2 |
0.234 |
0.543 |
| 41 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
0.228 |
0.117 |
| 42 |
253125_at |
AT4G36040
|
DJC23, DNA J protein C23, J11, DnaJ11 |
0.220 |
0.069 |
| 43 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.216 |
0.098 |
| 44 |
250738_at |
AT5G05730
|
AMT1, A-METHYL TRYPTOPHAN RESISTANT 1, ASA1, anthranilate synthase alpha subunit 1, JDL1, JASMONATE-INDUCED DEFECTIVE LATERAL ROOT 1, TRP5, TRYPTOPHAN BIOSYNTHESIS 5, WEI2, WEAK ETHYLENE INSENSITIVE 2 |
0.215 |
0.245 |
| 45 |
246981_at |
AT5G04840
|
[bZIP protein] |
0.199 |
0.155 |
| 46 |
265766_at |
AT2G48080
|
[oxidoreductase, 2OG-Fe(II) oxygenase family protein] |
0.197 |
0.113 |
| 47 |
260484_at |
AT1G68360
|
[C2H2 and C2HC zinc fingers superfamily protein] |
0.197 |
0.098 |
| 48 |
248886_at |
AT5G46110
|
APE2, ACCLIMATION OF PHOTOSYNTHESIS TO ENVIRONMENT 2, TPT, triose-phosphate ⁄ phosphate translocator |
-0.478 |
-0.234 |
| 49 |
255886_at |
AT1G20340
|
DRT112, DNA-DAMAGE-REPAIR/TOLERATION PROTEIN 112, PETE2, PLASTOCYANIN 2 |
-0.241 |
-0.270 |
| 50 |
259840_at |
AT1G52230
|
PSAH-2, PHOTOSYSTEM I SUBUNIT H-2, PSAH2, photosystem I subunit H2, PSI-H |
-0.238 |
-0.319 |
| 51 |
265033_at |
AT1G61520
|
LHCA3, photosystem I light harvesting complex gene 3 |
-0.229 |
-0.188 |
| 52 |
245044_at |
AT2G26500
|
[cytochrome b6f complex subunit (petM), putative] |
-0.229 |
-0.149 |
| 53 |
265287_at |
AT2G20260
|
PSAE-2, photosystem I subunit E-2 |
-0.224 |
-0.252 |
| 54 |
246200_at |
AT4G37240
|
unknown |
-0.222 |
0.021 |
| 55 |
258241_at |
AT3G27650
|
LBD25, LOB domain-containing protein 25 |
-0.220 |
0.122 |
| 56 |
258209_at |
AT3G14060
|
unknown |
-0.217 |
-0.038 |
| 57 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
-0.206 |
-0.237 |
| 58 |
265149_at |
AT1G51400
|
[Photosystem II 5 kD protein] |
-0.204 |
-0.313 |
| 59 |
261821_at |
AT1G11530
|
ATCXXS1, C-terminal cysteine residue is changed to a serine 1, CXXS1, C-terminal cysteine residue is changed to a serine 1 |
-0.190 |
-0.079 |
| 60 |
254970_at |
AT4G10340
|
LHCB5, light harvesting complex of photosystem II 5 |
-0.187 |
-0.317 |
| 61 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
-0.187 |
-0.011 |
| 62 |
262557_at |
AT1G31330
|
PSAF, photosystem I subunit F |
-0.186 |
-0.157 |
| 63 |
248273_at |
AT5G53500
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.183 |
-0.075 |
| 64 |
263736_at |
AT1G60000
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.171 |
-0.246 |
| 65 |
266551_at |
AT2G35260
|
unknown |
-0.168 |
-0.304 |
| 66 |
245586_at |
AT4G14980
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.168 |
0.081 |
| 67 |
265374_at |
AT2G06520
|
PSBX, photosystem II subunit X |
-0.165 |
-0.245 |
| 68 |
262916_at |
AT1G59700
|
ATGSTU16, glutathione S-transferase TAU 16, GSTU16, glutathione S-transferase TAU 16 |
-0.159 |
-0.410 |
| 69 |
251762_at |
AT3G55800
|
SBPASE, sedoheptulose-bisphosphatase |
-0.155 |
-0.424 |
| 70 |
260926_at |
AT1G21360
|
GLTP2, glycolipid transfer protein 2 |
-0.147 |
0.053 |
| 71 |
262632_at |
AT1G06680
|
OE23, OXYGEN EVOLVING COMPLEX SUBUNIT 23 KDA, OEE2, OXYGEN-EVOLVING ENHANCER PROTEIN 2, PSBP-1, photosystem II subunit P-1, PSII-P, PHOTOSYSTEM II SUBUNIT P |
-0.146 |
-0.233 |
| 72 |
249532_at |
AT5G38780
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.144 |
-0.082 |
| 73 |
259491_at |
AT1G15820
|
CP24, LHCB6, light harvesting complex photosystem II subunit 6 |
-0.142 |
-0.342 |
| 74 |
259316_at |
AT3G01175
|
unknown |
-0.140 |
0.220 |
| 75 |
256066_at |
AT1G06980
|
unknown |
-0.139 |
0.051 |
| 76 |
253936_at |
AT4G26880
|
[Stigma-specific Stig1 family protein] |
-0.139 |
0.087 |
| 77 |
264092_at |
AT1G79040
|
PSBR, photosystem II subunit R |
-0.139 |
-0.104 |
| 78 |
263114_at |
AT1G03130
|
PSAD-2, photosystem I subunit D-2 |
-0.132 |
-0.275 |
| 79 |
245454_at |
AT4G16920
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.129 |
0.045 |
| 80 |
263069_at |
AT2G17590
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.125 |
0.039 |
| 81 |
256336_at |
AT1G72030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.124 |
-0.463 |
| 82 |
265333_at |
AT2G18350
|
AtHB24, homeobox protein 24, HB24, homeobox protein 24, ZHD6, ZINC FINGER HOMEODOMAIN 6 |
-0.124 |
0.267 |
| 83 |
250158_at |
AT5G15190
|
unknown |
-0.115 |
0.215 |
| 84 |
263287_at |
AT2G36145
|
unknown |
-0.115 |
-0.316 |
| 85 |
250937_at |
AT5G03230
|
unknown |
-0.113 |
-0.027 |
| 86 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
-0.113 |
0.310 |
| 87 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
-0.110 |
-0.111 |
| 88 |
247073_at |
AT5G66570
|
MSP-1, MANGANESE-STABILIZING PROTEIN 1, OE33, OXYGEN EVOLVING COMPLEX 33 KILODALTON PROTEIN, OEE1, 33 KDA OXYGEN EVOLVING POLYPEPTIDE 1, OEE33, OXYGEN EVOLVING ENHANCER PROTEIN 33, PSBO-1, PS II OXYGEN-EVOLVING COMPLEX 1, PSBO1, PS II oxygen-evolving complex 1 |
-0.108 |
-0.280 |
| 89 |
251727_at |
AT3G56290
|
unknown |
-0.103 |
-0.155 |
| 90 |
260106_at |
AT1G35420
|
[alpha/beta-Hydrolases superfamily protein] |
-0.103 |
-0.305 |
| 91 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
-0.102 |
0.135 |
| 92 |
258025_at |
AT3G19480
|
3-PGDH, PGDH3, phosphoglycerate dehydrogenase 3 |
-0.100 |
-0.283 |
| 93 |
260887_at |
AT1G29160
|
[Dof-type zinc finger DNA-binding family protein] |
-0.099 |
0.328 |
| 94 |
254102_at |
AT4G25050
|
ACP4, acyl carrier protein 4 |
-0.094 |
-0.284 |