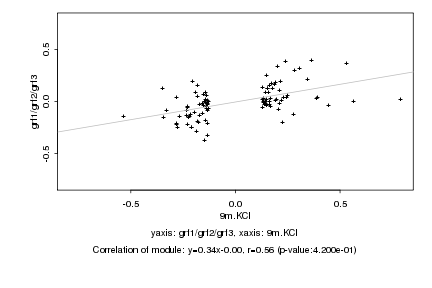
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
9m.KCl |
Log2 signal ratio
grf1/grf2/grf3 |
| 1 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.787 |
0.024 |
| 2 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
0.564 |
0.001 |
| 3 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.529 |
0.368 |
| 4 |
262124_at |
AT1G59660
|
[Nucleoporin autopeptidase] |
0.443 |
-0.036 |
| 5 |
264912_at |
AT1G60750
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.390 |
0.044 |
| 6 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
0.383 |
0.036 |
| 7 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.363 |
0.401 |
| 8 |
254286_at |
AT4G22950
|
AGL19, AGAMOUS-like 19, GL19 |
0.340 |
0.219 |
| 9 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.303 |
0.325 |
| 10 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.278 |
0.301 |
| 11 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.273 |
-0.124 |
| 12 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.248 |
0.065 |
| 13 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.240 |
0.046 |
| 14 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.237 |
0.386 |
| 15 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.225 |
0.046 |
| 16 |
264647_at |
AT1G09090
|
ATRBOHB, respiratory burst oxidase homolog B, ATRBOHB-BETA, RBOHB, respiratory burst oxidase homolog B |
0.221 |
-0.200 |
| 17 |
247145_at |
AT5G65600
|
LecRK-IX.2, L-type lectin receptor kinase IX.2 |
0.219 |
0.014 |
| 18 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.211 |
0.198 |
| 19 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.209 |
-0.010 |
| 20 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.207 |
0.112 |
| 21 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.204 |
-0.075 |
| 22 |
265062_at |
AT1G61550
|
[S-locus lectin protein kinase family protein] |
0.197 |
0.341 |
| 23 |
257547_at |
AT3G13000
|
unknown |
0.192 |
0.022 |
| 24 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.188 |
0.191 |
| 25 |
250499_at |
AT5G09730
|
ATBX3, ATBXL3, BETA-XYLOSIDASE 3, BX3, BXL3, beta-xylosidase 3, XYL3 |
0.187 |
0.017 |
| 26 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
0.185 |
0.164 |
| 27 |
253573_at |
AT4G31020
|
[alpha/beta-Hydrolases superfamily protein] |
0.175 |
0.132 |
| 28 |
266269_at |
AT2G29480
|
ATGSTU2, glutathione S-transferase tau 2, GST20, GLUTATHIONE S-TRANSFERASE 20, GSTU2, glutathione S-transferase tau 2 |
0.169 |
0.177 |
| 29 |
261117_at |
AT1G75310
|
AUL1, auxilin-like 1 |
0.166 |
0.036 |
| 30 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.164 |
-0.046 |
| 31 |
247855_at |
AT5G58210
|
[hydroxyproline-rich glycoprotein family protein] |
0.161 |
-0.026 |
| 32 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
0.159 |
0.004 |
| 33 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.158 |
0.163 |
| 34 |
267300_at |
AT2G30140
|
UGT87A2, UDP-glucosyl transferase 87A2 |
0.155 |
0.092 |
| 35 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
0.150 |
0.131 |
| 36 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
0.150 |
0.127 |
| 37 |
258351_at |
AT3G17700
|
ATCNGC20, CYCLIC NUCLEOTIDE-GATED CHANNEL 20, CNBT1, cyclic nucleotide-binding transporter 1, CNGC20, cyclic nucleotide gated channel 20 |
0.148 |
0.027 |
| 38 |
246055_at |
AT5G08380
|
AGAL1, alpha-galactosidase 1, AtAGAL1, alpha-galactosidase 1 |
0.148 |
-0.032 |
| 39 |
262362_at |
AT1G72840
|
[Disease resistance protein (TIR-NBS-LRR class)] |
0.147 |
-0.024 |
| 40 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.146 |
0.250 |
| 41 |
261460_at |
AT1G07880
|
ATMPK13 |
0.143 |
0.095 |
| 42 |
256010_at |
AT1G19220
|
ARF11, AUXIN RESPONSE FACTOR11, ARF19, auxin response factor 19, IAA22, indole-3-acetic acid inducible 22 |
0.141 |
0.001 |
| 43 |
265659_at |
AT2G25440
|
AtRLP20, receptor like protein 20, RLP20, receptor like protein 20 |
0.140 |
-0.013 |
| 44 |
266325_at |
AT2G46630
|
unknown |
0.138 |
-0.021 |
| 45 |
253258_at |
AT4G34400
|
[AP2/B3-like transcriptional factor family protein] |
0.137 |
-0.009 |
| 46 |
258108_at |
AT3G23570
|
[alpha/beta-Hydrolases superfamily protein] |
0.133 |
-0.006 |
| 47 |
254944_at |
AT4G10930
|
unknown |
0.130 |
0.004 |
| 48 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.130 |
0.035 |
| 49 |
267140_at |
AT2G38250
|
[Homeodomain-like superfamily protein] |
0.129 |
0.025 |
| 50 |
256394_at |
AT3G06290
|
AtSAC3B, SAC3B, yeast Sac3 homolog B |
0.128 |
-0.048 |
| 51 |
256081_at |
AT1G20700
|
ATWOX14, WUSCHEL RELATED HOMEOBOX 14, WOX14, WUSCHEL related homeobox 14 |
0.127 |
0.141 |
| 52 |
248886_at |
AT5G46110
|
APE2, ACCLIMATION OF PHOTOSYNTHESIS TO ENVIRONMENT 2, TPT, triose-phosphate ⁄ phosphate translocator |
-0.539 |
-0.136 |
| 53 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-0.351 |
0.133 |
| 54 |
247320_at |
AT5G64040
|
PSAN |
-0.346 |
-0.144 |
| 55 |
266191_at |
AT2G39040
|
[Peroxidase superfamily protein] |
-0.332 |
-0.082 |
| 56 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.286 |
0.040 |
| 57 |
248151_at |
AT5G54270
|
LHCB3, light-harvesting chlorophyll B-binding protein 3, LHCB3*1 |
-0.285 |
-0.210 |
| 58 |
255934_at |
AT1G12740
|
CYP87A2, cytochrome P450, family 87, subfamily A, polypeptide 2 |
-0.285 |
-0.216 |
| 59 |
255886_at |
AT1G20340
|
DRT112, DNA-DAMAGE-REPAIR/TOLERATION PROTEIN 112, PETE2, PLASTOCYANIN 2 |
-0.279 |
-0.247 |
| 60 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
-0.268 |
-0.137 |
| 61 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
-0.235 |
-0.081 |
| 62 |
248408_at |
AT5G51520
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.234 |
-0.128 |
| 63 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.233 |
-0.056 |
| 64 |
258285_at |
AT3G16140
|
PSAH-1, photosystem I subunit H-1 |
-0.231 |
-0.214 |
| 65 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.231 |
-0.047 |
| 66 |
245674_at |
AT1G56680
|
[Chitinase family protein] |
-0.227 |
-0.145 |
| 67 |
264092_at |
AT1G79040
|
PSBR, photosystem II subunit R |
-0.224 |
-0.135 |
| 68 |
245399_at |
AT4G17340
|
DELTA-TIP2, TIP2;2, tonoplast intrinsic protein 2;2 |
-0.218 |
-0.124 |
| 69 |
254970_at |
AT4G10340
|
LHCB5, light harvesting complex of photosystem II 5 |
-0.214 |
-0.243 |
| 70 |
262557_at |
AT1G31330
|
PSAF, photosystem I subunit F |
-0.212 |
-0.240 |
| 71 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.208 |
0.194 |
| 72 |
250165_at |
AT5G15290
|
CASP5, Casparian strip membrane domain protein 5 |
-0.200 |
-0.099 |
| 73 |
258209_at |
AT3G14060
|
unknown |
-0.191 |
0.094 |
| 74 |
266636_at |
AT2G35370
|
GDCH, glycine decarboxylase complex H |
-0.190 |
-0.285 |
| 75 |
251814_at |
AT3G54890
|
LHCA1, photosystem I light harvesting complex gene 1 |
-0.186 |
-0.184 |
| 76 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-0.183 |
0.163 |
| 77 |
252115_at |
AT3G51600
|
LTP5, lipid transfer protein 5 |
-0.182 |
0.051 |
| 78 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.180 |
-0.199 |
| 79 |
252834_at |
AT4G40070
|
[RING/U-box superfamily protein] |
-0.176 |
-0.128 |
| 80 |
245990_at |
AT5G20640
|
unknown |
-0.175 |
-0.028 |
| 81 |
252430_at |
AT3G47470
|
CAB4, LHCA4, light-harvesting chlorophyll-protein complex I subunit A4 |
-0.160 |
-0.112 |
| 82 |
256714_at |
AT2G34080
|
[Cysteine proteinases superfamily protein] |
-0.160 |
-0.011 |
| 83 |
253155_at |
AT4G35720
|
unknown |
-0.154 |
0.070 |
| 84 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
-0.153 |
-0.037 |
| 85 |
255129_at |
AT4G08290
|
UMAMIT20, Usually multiple acids move in and out Transporters 20 |
-0.152 |
-0.368 |
| 86 |
261091_at |
AT1G07550
|
[Leucine-rich repeat protein kinase family protein] |
-0.150 |
0.009 |
| 87 |
261931_at |
AT1G22430
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.145 |
-0.182 |
| 88 |
255942_at |
AT1G22360
|
AtUGT85A2, UDP-glucosyl transferase 85A2, UGT85A2, UDP-glucosyl transferase 85A2 |
-0.145 |
0.000 |
| 89 |
249532_at |
AT5G38780
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.144 |
0.028 |
| 90 |
261647_at |
AT1G27740
|
RSL4, root hair defective 6-like 4 |
-0.144 |
0.087 |
| 91 |
258241_at |
AT3G27650
|
LBD25, LOB domain-containing protein 25 |
-0.143 |
0.058 |
| 92 |
252536_at |
AT3G45700
|
[Major facilitator superfamily protein] |
-0.143 |
-0.047 |
| 93 |
267085_at |
AT2G32620
|
ATCSLB02, cellulose synthase-like B, ATCSLB2, CELLULOSE SYNTHASE LIKE B2, CSLB02, CELLULOSE SYNTHASE LIKE B2, CSLB02, cellulose synthase-like B |
-0.140 |
-0.061 |
| 94 |
251244_at |
AT3G62060
|
[Pectinacetylesterase family protein] |
-0.138 |
0.016 |
| 95 |
250479_at |
AT5G10260
|
AtRABH1e, RAB GTPase homolog H1E, RABH1e, RAB GTPase homolog H1E |
-0.137 |
-0.014 |
| 96 |
246390_at |
AT1G77330
|
ACO5, ACC oxidase 5 |
-0.137 |
-0.081 |
| 97 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.136 |
-0.317 |
| 98 |
259840_at |
AT1G52230
|
PSAH-2, PHOTOSYSTEM I SUBUNIT H-2, PSAH2, photosystem I subunit H2, PSI-H |
-0.134 |
-0.209 |
| 99 |
254914_at |
AT4G11290
|
[Peroxidase superfamily protein] |
-0.134 |
-0.024 |
| 100 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.133 |
-0.060 |
| 101 |
250128_at |
AT5G16540
|
ZFN3, zinc finger nuclease 3 |
-0.133 |
0.007 |