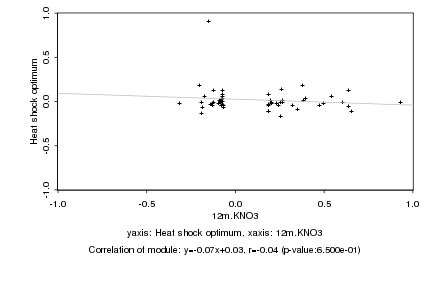
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
12m.KNO3 |
Log2 signal ratio
Heat shock optimum |
| 1 |
257645_at |
AT3G25790
|
[myb-like transcription factor family protein] |
0.928 |
0.000 |
| 2 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
0.653 |
-0.111 |
| 3 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.637 |
0.135 |
| 4 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.637 |
-0.053 |
| 5 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
0.602 |
-0.002 |
| 6 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.538 |
0.062 |
| 7 |
252220_at |
AT3G49940
|
LBD38, LOB domain-containing protein 38 |
0.496 |
-0.013 |
| 8 |
255734_at |
AT1G25550
|
[myb-like transcription factor family protein] |
0.471 |
-0.035 |
| 9 |
260522_x_at |
AT2G41730
|
unknown |
0.393 |
0.042 |
| 10 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
0.383 |
0.015 |
| 11 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.374 |
0.190 |
| 12 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
0.345 |
-0.082 |
| 13 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.319 |
-0.035 |
| 14 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
0.263 |
-0.000 |
| 15 |
259961_at |
AT1G53700
|
PK3AT, PROTEIN KINASE 3 ARABIDOPSIS THALIANA, WAG1, WAG 1 |
0.260 |
0.020 |
| 16 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.258 |
0.140 |
| 17 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
0.251 |
-0.160 |
| 18 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
0.249 |
0.000 |
| 19 |
249325_at |
AT5G40850
|
UPM1, urophorphyrin methylase 1 |
0.239 |
-0.044 |
| 20 |
247674_at |
AT5G59930
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.229 |
-0.013 |
| 21 |
257975_at |
AT3G20830
|
AGC2-4, AGC2 kinase 4, UCNL, UNICORN-like |
0.200 |
-0.016 |
| 22 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
0.199 |
-0.011 |
| 23 |
255230_at |
AT4G05390
|
ATRFNR1, root FNR 1, RFNR1, root FNR 1 |
0.192 |
0.019 |
| 24 |
247199_at |
AT5G65210
|
TGA1, TGACG sequence-specific binding protein 1 |
0.185 |
-0.034 |
| 25 |
250914_at |
AT5G03780
|
TRFL10, TRF-like 10 |
0.184 |
-0.032 |
| 26 |
248199_at |
AT5G54170
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.183 |
0.086 |
| 27 |
258350_at |
AT3G17510
|
CIPK1, CBL-interacting protein kinase 1, SnRK3.16, SNF1-RELATED PROTEIN KINASE 3.16 |
0.183 |
-0.107 |
| 28 |
260484_at |
AT1G68360
|
[C2H2 and C2HC zinc fingers superfamily protein] |
0.181 |
-0.104 |
| 29 |
248886_at |
AT5G46110
|
APE2, ACCLIMATION OF PHOTOSYNTHESIS TO ENVIRONMENT 2, TPT, triose-phosphate ⁄ phosphate translocator |
-0.317 |
-0.014 |
| 30 |
246395_at |
AT1G58170
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.203 |
0.184 |
| 31 |
245044_at |
AT2G26500
|
[cytochrome b6f complex subunit (petM), putative] |
-0.196 |
-0.008 |
| 32 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
-0.194 |
-0.135 |
| 33 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
-0.187 |
-0.063 |
| 34 |
265083_at |
AT1G03820
|
unknown |
-0.178 |
0.061 |
| 35 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
-0.154 |
0.911 |
| 36 |
246518_at |
AT5G15770
|
AtGNA1, glucose-6-phosphate acetyltransferase 1, GNA1, glucose-6-phosphate acetyltransferase 1 |
-0.146 |
-0.029 |
| 37 |
249601_at |
AT5G37980
|
[Zinc-binding dehydrogenase family protein] |
-0.134 |
-0.015 |
| 38 |
257593_at |
AT3G24840
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.131 |
-0.035 |
| 39 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
-0.128 |
0.130 |
| 40 |
263232_at |
AT1G05700
|
[Leucine-rich repeat transmembrane protein kinase protein] |
-0.125 |
-0.007 |
| 41 |
254748_at |
AT4G13120
|
[hAT-like transposase family (hobo/Ac/Tam3), has a 2.6e-50 P-value blast match to GB:AAD24567 transposase Tag2 (hAT-element) (Arabidopsis thaliana)] |
-0.097 |
-0.020 |
| 42 |
262338_at |
AT1G64185
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
-0.093 |
0.015 |
| 43 |
245674_at |
AT1G56680
|
[Chitinase family protein] |
-0.089 |
0.001 |
| 44 |
259491_at |
AT1G15820
|
CP24, LHCB6, light harvesting complex photosystem II subunit 6 |
-0.085 |
-0.004 |
| 45 |
252202_at |
AT3G50300
|
[HXXXD-type acyl-transferase family protein] |
-0.084 |
-0.038 |
| 46 |
265044_at |
AT1G03950
|
VPS2.3, vacuolar protein sorting-associated protein 2.3 |
-0.084 |
-0.036 |
| 47 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.079 |
0.034 |
| 48 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
-0.078 |
0.131 |
| 49 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
-0.077 |
0.060 |
| 50 |
264259_at |
AT1G09290
|
unknown |
-0.074 |
0.003 |
| 51 |
251281_at |
AT3G61640
|
AGP20, arabinogalactan protein 20, AtAGP20, Arabinogalactan protein 20 |
-0.074 |
0.083 |
| 52 |
267075_at |
AT2G41070
|
ATBZIP12, DPBF4, EEL, ENHANCED EM LEVEL |
-0.073 |
0.037 |
| 53 |
262568_at |
AT1G34290
|
AtRLP5, receptor like protein 5, RLP5, receptor like protein 5 |
-0.073 |
-0.025 |
| 54 |
259321_at |
AT3G01040
|
GAUT13, galacturonosyltransferase 13 |
-0.072 |
-0.060 |
| 55 |
265787_at |
AT2G07320
|
unknown |
-0.070 |
-0.038 |