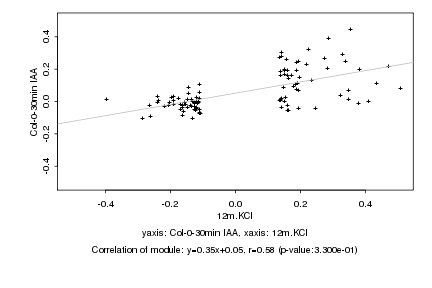
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
12m.KCl |
Log2 signal ratio
Col-0-30min IAA |
| 1 |
250446_at |
AT5G10770
|
[Eukaryotic aspartyl protease family protein] |
0.507 |
0.086 |
| 2 |
253643_at |
AT4G29780
|
unknown |
0.469 |
0.221 |
| 3 |
260522_x_at |
AT2G41730
|
unknown |
0.434 |
0.116 |
| 4 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
0.409 |
0.005 |
| 5 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.381 |
0.202 |
| 6 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
0.377 |
-0.012 |
| 7 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.353 |
0.447 |
| 8 |
254743_at |
AT4G13420
|
ATHAK5, ARABIDOPSIS THALIANA HIGH AFFINITY K+ TRANSPORTER 5, HAK5, high affinity K+ transporter 5 |
0.348 |
0.013 |
| 9 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.346 |
0.070 |
| 10 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.337 |
0.252 |
| 11 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.328 |
0.293 |
| 12 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.322 |
0.042 |
| 13 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.285 |
0.395 |
| 14 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.282 |
0.209 |
| 15 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.272 |
0.272 |
| 16 |
254098_at |
AT4G25100
|
ATFSD1, ARABIDOPSIS FE SUPEROXIDE DISMUTASE 1, FSD1, Fe superoxide dismutase 1 |
0.246 |
-0.037 |
| 17 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.232 |
0.136 |
| 18 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.222 |
0.325 |
| 19 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.218 |
0.234 |
| 20 |
259979_at |
AT1G76600
|
unknown |
0.196 |
0.152 |
| 21 |
256306_at |
AT1G30370
|
DLAH, DAD1-like acylhydrolase |
0.194 |
0.253 |
| 22 |
245017_at |
ATCG00510
|
PSAI, photsystem I subunit I |
0.193 |
-0.037 |
| 23 |
250745_at |
AT5G05850
|
PIRL1, plant intracellular ras group-related LRR 1 |
0.192 |
0.072 |
| 24 |
261545_at |
AT1G63530
|
unknown |
0.191 |
0.113 |
| 25 |
263931_at |
AT2G36220
|
unknown |
0.185 |
0.243 |
| 26 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
0.185 |
0.197 |
| 27 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
0.185 |
0.076 |
| 28 |
263809_at |
AT2G04570
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.183 |
0.110 |
| 29 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.177 |
0.099 |
| 30 |
248981_at |
AT5G45110
|
ATNPR3, NPR3, NPR1-like protein 3 |
0.172 |
0.162 |
| 31 |
265567_at |
AT2G05580
|
[Glycine-rich protein family] |
0.163 |
-0.053 |
| 32 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.161 |
0.145 |
| 33 |
259137_at |
AT3G10300
|
[Calcium-binding EF-hand family protein] |
0.160 |
0.166 |
| 34 |
245550_at |
AT4G15330
|
CYP705A1, cytochrome P450, family 705, subfamily A, polypeptide 1 |
0.159 |
-0.055 |
| 35 |
249746_at |
AT5G24590
|
ANAC091, Arabidopsis NAC domain containing protein 91, TIP, TCV-interacting protein |
0.159 |
0.197 |
| 36 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
0.159 |
-0.021 |
| 37 |
262801_at |
AT1G21010
|
unknown |
0.155 |
0.263 |
| 38 |
252539_at |
AT3G45730
|
unknown |
0.153 |
0.026 |
| 39 |
267069_at |
AT2G41010
|
ATCAMBP25, calmodulin (CAM)-binding protein of 25 kDa, CAMBP25, calmodulin (CAM)-binding protein of 25 kDa |
0.150 |
0.202 |
| 40 |
257751_at |
AT3G18690
|
MKS1, MAP kinase substrate 1 |
0.149 |
0.194 |
| 41 |
266124_at |
AT2G45080
|
cycp3;1, cyclin p3;1 |
0.148 |
0.168 |
| 42 |
246055_at |
AT5G08380
|
AGAL1, alpha-galactosidase 1, AtAGAL1, alpha-galactosidase 1 |
0.148 |
0.002 |
| 43 |
249208_at |
AT5G42650
|
AOS, allene oxide synthase, CYP74A, CYTOCHROME P450 74A, DDE2, DELAYED DEHISCENCE 2 |
0.145 |
0.087 |
| 44 |
247808_at |
AT5G58190
|
ECT10, evolutionarily conserved C-terminal region 10 |
0.141 |
-0.031 |
| 45 |
256763_at |
AT3G16860
|
COBL8, COBRA-like protein 8 precursor |
0.140 |
0.306 |
| 46 |
261476_at |
AT1G14480
|
[Ankyrin repeat family protein] |
0.139 |
0.279 |
| 47 |
251157_at |
AT3G63140
|
CSP41A, chloroplast stem-loop binding protein of 41 kDa |
0.139 |
0.023 |
| 48 |
258253_at |
AT3G26760
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.138 |
0.162 |
| 49 |
261441_at |
AT1G28470
|
ANAC010, NAC domain containing protein 10, NAC010, NAC domain containing protein 10, SND3, SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 3 |
0.137 |
0.008 |
| 50 |
258436_at |
AT3G16720
|
ATL2, TOXICOS EN LEVADURA 2, TL2, TOXICOS EN LEVADURA 2 |
0.136 |
0.188 |
| 51 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
0.136 |
0.015 |
| 52 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.134 |
0.274 |
| 53 |
258733_at |
AT3G05850
|
MUG7, MUSTANG 7 |
0.133 |
0.010 |
| 54 |
255886_at |
AT1G20340
|
DRT112, DNA-DAMAGE-REPAIR/TOLERATION PROTEIN 112, PETE2, PLASTOCYANIN 2 |
-0.400 |
0.015 |
| 55 |
258341_at |
AT3G22790
|
NET1A, Networked 1A |
-0.289 |
-0.101 |
| 56 |
258033_at |
AT3G21250
|
ABCC8, ATP-binding cassette C8, ATMRP6, ARABIDOPSIS THALIANA MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 6, MRP6, multidrug resistance-associated protein 6 |
-0.267 |
-0.022 |
| 57 |
256131_at |
AT1G13600
|
AtbZIP58, basic leucine-zipper 58, bZIP58, basic leucine-zipper 58 |
-0.262 |
-0.089 |
| 58 |
253738_at |
AT4G28750
|
PSAE-1, PSA E1 KNOCKOUT |
-0.242 |
-0.001 |
| 59 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-0.242 |
0.035 |
| 60 |
256309_at |
AT1G30380
|
PSAK, photosystem I subunit K |
-0.238 |
0.010 |
| 61 |
245044_at |
AT2G26500
|
[cytochrome b6f complex subunit (petM), putative] |
-0.220 |
-0.030 |
| 62 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
-0.207 |
-0.022 |
| 63 |
251664_at |
AT3G56940
|
ACSF, CHL27, CRD1, COPPER RESPONSE DEFECT 1 |
-0.206 |
0.000 |
| 64 |
255904_at |
AT1G17860
|
[Kunitz family trypsin and protease inhibitor protein] |
-0.198 |
0.029 |
| 65 |
261031_at |
AT1G17360
|
unknown |
-0.193 |
0.037 |
| 66 |
250550_at |
AT5G07870
|
[HXXXD-type acyl-transferase family protein] |
-0.192 |
-0.015 |
| 67 |
253936_at |
AT4G26880
|
[Stigma-specific Stig1 family protein] |
-0.191 |
0.010 |
| 68 |
263232_at |
AT1G05700
|
[Leucine-rich repeat transmembrane protein kinase protein] |
-0.176 |
0.024 |
| 69 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
-0.170 |
-0.048 |
| 70 |
246375_at |
AT1G51830
|
[Leucine-rich repeat protein kinase family protein] |
-0.170 |
-0.016 |
| 71 |
265471_at |
AT2G37130
|
[Peroxidase superfamily protein] |
-0.166 |
-0.020 |
| 72 |
259699_at |
AT1G68940
|
[Armadillo/beta-catenin-like repeat family protein] |
-0.164 |
-0.083 |
| 73 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
-0.164 |
-0.035 |
| 74 |
263736_at |
AT1G60000
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.164 |
-0.024 |
| 75 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.162 |
-0.058 |
| 76 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.159 |
-0.006 |
| 77 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.154 |
-0.017 |
| 78 |
263470_at |
AT2G31900
|
ATMYO5, MYOSIN 5, ATXIF, MYOSIN XI F, XIF, myosin-like protein XIF |
-0.150 |
-0.035 |
| 79 |
249360_at |
AT5G40530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.149 |
0.016 |
| 80 |
254659_at |
AT4G18240
|
ATSS4, ARABIDOPSIS THALIANA STARCH SYNTHASE 4, SS4, starch synthase 4, SSIV, STARCH SYNTHASE 4 |
-0.146 |
0.050 |
| 81 |
254586_at |
AT4G19510
|
[Disease resistance protein (TIR-NBS-LRR class)] |
-0.146 |
0.087 |
| 82 |
254939_at |
AT4G10800
|
unknown |
-0.139 |
-0.021 |
| 83 |
256652_at |
AT3G18850
|
LPAT5, lysophosphatidyl acyltransferase 5 |
-0.137 |
-0.030 |
| 84 |
265374_at |
AT2G06520
|
PSBX, photosystem II subunit X |
-0.136 |
0.016 |
| 85 |
263805_at |
AT2G40400
|
unknown |
-0.135 |
0.006 |
| 86 |
245990_at |
AT5G20640
|
unknown |
-0.134 |
-0.102 |
| 87 |
248427_at |
AT5G51750
|
ATSBT1.3, subtilase 1.3, SBT1.3, subtilase 1.3 |
-0.129 |
-0.011 |
| 88 |
250243_at |
AT5G13630
|
ABAR, ABA-BINDING PROTEIN, CCH, CONDITIONAL CHLORINA, CCH1, CHLH, H SUBUNIT OF MG-CHELATASE, GUN5, GENOMES UNCOUPLED 5 |
-0.129 |
-0.026 |
| 89 |
265814_at |
AT2G17930
|
[Phosphatidylinositol 3- and 4-kinase family protein with FAT domain] |
-0.128 |
-0.047 |
| 90 |
266469_at |
AT2G31180
|
ATMYB14, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 14, MYB14, myb domain protein 14, MYB14AT |
-0.127 |
-0.005 |
| 91 |
246205_at |
AT4G36970
|
[Remorin family protein] |
-0.124 |
-0.055 |
| 92 |
254851_at |
AT4G12010
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.122 |
0.006 |
| 93 |
260367_at |
AT1G69760
|
unknown |
-0.120 |
-0.036 |
| 94 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
-0.120 |
0.031 |
| 95 |
263607_at |
AT2G16270
|
unknown |
-0.120 |
-0.039 |
| 96 |
263122_at |
AT1G78510
|
AtSPS1, SPS1, solanesyl diphosphate synthase 1 |
-0.117 |
-0.008 |
| 97 |
257831_at |
AT3G26710
|
CCB1, cofactor assembly of complex C |
-0.115 |
0.005 |
| 98 |
255501_at |
AT4G02400
|
[U3 ribonucleoprotein (Utp) family protein] |
-0.115 |
-0.005 |
| 99 |
263436_at |
AT2G28690
|
unknown |
-0.113 |
-0.067 |
| 100 |
263909_at |
AT2G36490
|
AtROS1, DML1, demeter-like 1, ROS1, REPRESSOR OF SILENCING1 |
-0.113 |
-0.069 |
| 101 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-0.113 |
0.106 |
| 102 |
262891_at |
AT1G79460
|
ATKS, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE, ATKS1, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE 1, GA2, GA REQUIRING 2, KS, KS1, ENT-KAURENE SYNTHASE 1 |
-0.112 |
0.021 |
| 103 |
261557_at |
AT1G63640
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
-0.112 |
-0.046 |
| 104 |
247399_at |
AT5G62960
|
unknown |
-0.111 |
0.057 |
| 105 |
247310_at |
AT5G63950
|
CHR24, chromatin remodeling 24 |
-0.110 |
-0.070 |