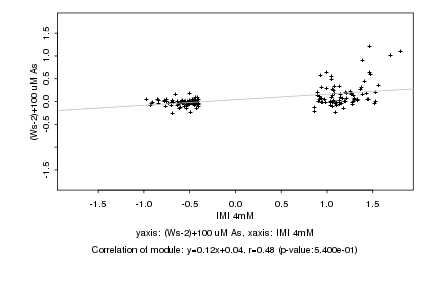
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
IMI 4mM |
Log2 signal ratio
(Ws-2)+100 uM As |
| 1 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
1.799 |
1.112 |
| 2 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
1.689 |
1.020 |
| 3 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
1.554 |
0.352 |
| 4 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
1.526 |
0.019 |
| 5 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
1.522 |
0.207 |
| 6 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
1.516 |
-0.035 |
| 7 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
1.468 |
0.603 |
| 8 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
1.464 |
1.217 |
| 9 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
1.457 |
0.637 |
| 10 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
1.448 |
0.051 |
| 11 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
1.434 |
0.047 |
| 12 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
1.428 |
0.181 |
| 13 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
1.406 |
0.457 |
| 14 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
1.388 |
0.174 |
| 15 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
1.384 |
0.905 |
| 16 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
1.375 |
0.308 |
| 17 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
1.359 |
0.277 |
| 18 |
246197_at |
AT4G37010
|
CEN2, centrin 2 |
1.331 |
0.048 |
| 19 |
256593_at |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
1.327 |
0.037 |
| 20 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
1.296 |
0.084 |
| 21 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
1.290 |
0.054 |
| 22 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
1.289 |
0.151 |
| 23 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
1.287 |
0.007 |
| 24 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
1.275 |
-0.013 |
| 25 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
1.273 |
-0.048 |
| 26 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
1.264 |
0.160 |
| 27 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
1.263 |
0.189 |
| 28 |
256407_at |
AT1G66570
|
ATSUC7, sucrose-proton symporter 7, SUC7, sucrose-proton symporter 7 |
1.250 |
0.238 |
| 29 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
1.210 |
0.069 |
| 30 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
1.208 |
0.176 |
| 31 |
254001_at |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
1.201 |
0.219 |
| 32 |
249454_at |
AT5G39520
|
unknown |
1.196 |
0.014 |
| 33 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
1.182 |
0.019 |
| 34 |
267178_at |
AT2G37750
|
unknown |
1.174 |
-0.145 |
| 35 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
1.163 |
0.071 |
| 36 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
1.150 |
-0.026 |
| 37 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
1.145 |
0.109 |
| 38 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
1.139 |
0.157 |
| 39 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
1.134 |
0.337 |
| 40 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
1.131 |
-0.050 |
| 41 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
1.127 |
-0.005 |
| 42 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.126 |
0.038 |
| 43 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
1.100 |
-0.014 |
| 44 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
1.097 |
-0.071 |
| 45 |
266376_at |
AT2G14620
|
XTH10, xyloglucan endotransglucosylase/hydrolase 10 |
1.089 |
-0.032 |
| 46 |
260744_at |
AT1G15010
|
unknown |
1.083 |
-0.219 |
| 47 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
1.074 |
0.192 |
| 48 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
1.073 |
0.335 |
| 49 |
261545_at |
AT1G63530
|
unknown |
1.071 |
0.003 |
| 50 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
1.068 |
0.250 |
| 51 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
1.068 |
0.026 |
| 52 |
248528_at |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
1.058 |
-0.103 |
| 53 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
1.057 |
0.285 |
| 54 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
1.055 |
0.133 |
| 55 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
1.053 |
-0.001 |
| 56 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
1.045 |
0.504 |
| 57 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
1.043 |
0.552 |
| 58 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
1.040 |
0.093 |
| 59 |
246133_at |
AT5G20960
|
AAO1, aldehyde oxidase 1, AO1, aldehyde oxidase 1, AOalpha, aldehyde oxidase alpha, AT-AO1, ARABIDOPSIS THALIANA ALDEHYDE OXIDASE 1, ATAO, AtAO1, Arabidopsis thaliana aldehyde oxidase 1 |
1.038 |
-0.010 |
| 60 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
1.036 |
-0.086 |
| 61 |
264648_at |
AT1G09080
|
BIP3, binding protein 3 |
1.031 |
0.011 |
| 62 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
0.994 |
0.292 |
| 63 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
0.992 |
0.643 |
| 64 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.974 |
-0.013 |
| 65 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
0.967 |
0.054 |
| 66 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.947 |
-0.019 |
| 67 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.937 |
0.310 |
| 68 |
251743_at |
AT3G55890
|
[Yippee family putative zinc-binding protein] |
0.937 |
0.078 |
| 69 |
257591_at |
AT3G24900
|
AtRLP39, receptor like protein 39, RLP39, receptor like protein 39 |
0.931 |
-0.003 |
| 70 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.928 |
0.572 |
| 71 |
254453_at |
AT4G21120
|
AAT1, amino acid transporter 1, CAT1, CATIONIC AMINO ACID TRANSPORTER 1 |
0.925 |
0.074 |
| 72 |
259150_at |
AT3G10320
|
[Glycosyltransferase family 61 protein] |
0.914 |
0.127 |
| 73 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.912 |
0.047 |
| 74 |
246373_at |
AT1G51860
|
[Leucine-rich repeat protein kinase family protein] |
0.897 |
0.008 |
| 75 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.895 |
0.200 |
| 76 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.892 |
0.147 |
| 77 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
0.859 |
-0.122 |
| 78 |
263851_at |
AT2G04460
|
unknown |
0.855 |
-0.218 |
| 79 |
254122_at |
AT4G24510
|
CER2, ECERIFERUM 2, VC-2, VC2 |
-0.982 |
0.052 |
| 80 |
259660_at |
AT1G55260
|
LTPG6, glycosylphosphatidylinositol-anchored lipid protein transfer 6 |
-0.931 |
-0.079 |
| 81 |
245688_at |
AT1G28290
|
AGP31, arabinogalactan protein 31 |
-0.922 |
-0.025 |
| 82 |
263431_at |
AT2G22170
|
PLAT2, PLAT domain protein 2 |
-0.917 |
-0.004 |
| 83 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.856 |
0.054 |
| 84 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.850 |
0.032 |
| 85 |
264987_at |
AT1G27030
|
unknown |
-0.849 |
-0.024 |
| 86 |
249410_at |
AT5G40380
|
CRK42, cysteine-rich RLK (RECEPTOR-like protein kinase) 42 |
-0.781 |
0.031 |
| 87 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
-0.776 |
0.016 |
| 88 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
-0.767 |
-0.096 |
| 89 |
255690_at |
AT4G00360
|
ATT1, ABERRANT INDUCTION OF TYPE THREE 1, CYP86A2, cytochrome P450, family 86, subfamily A, polypeptide 2 |
-0.759 |
0.018 |
| 90 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
-0.757 |
0.045 |
| 91 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.733 |
-0.036 |
| 92 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.706 |
-0.017 |
| 93 |
258470_at |
AT3G06035
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.701 |
-0.080 |
| 94 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.695 |
-0.249 |
| 95 |
250261_at |
AT5G13400
|
[Major facilitator superfamily protein] |
-0.688 |
0.025 |
| 96 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
-0.686 |
-0.001 |
| 97 |
261570_at |
AT1G01120
|
KCS1, 3-ketoacyl-CoA synthase 1 |
-0.658 |
0.161 |
| 98 |
261480_at |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-0.643 |
-0.066 |
| 99 |
245743_at |
AT1G51080
|
unknown |
-0.641 |
0.002 |
| 100 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
-0.631 |
-0.042 |
| 101 |
253286_at |
AT4G34260
|
AXY8, ALTERED XYLOGLUCAN 8, FUC95A |
-0.623 |
-0.053 |
| 102 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
-0.614 |
-0.145 |
| 103 |
259788_at |
AT1G29670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.609 |
-0.016 |
| 104 |
263574_at |
AT2G16990
|
[Major facilitator superfamily protein] |
-0.602 |
-0.116 |
| 105 |
247288_at |
AT5G64330
|
JK218, NPH3, NON-PHOTOTROPIC HYPOCOTYL 3, RPT3, ROOT PHOTOTROPISM 3 |
-0.602 |
-0.019 |
| 106 |
253493_at |
AT4G31820
|
ENP, ENHANCER OF PINOID, MAB4, MACCHI-BOU 4, NPY1, NAKED PINS IN YUC MUTANTS 1 |
-0.599 |
0.011 |
| 107 |
253090_at |
AT4G36360
|
BGAL3, beta-galactosidase 3 |
-0.587 |
0.012 |
| 108 |
257017_at |
AT3G19620
|
[Glycosyl hydrolase family protein] |
-0.581 |
-0.035 |
| 109 |
248969_at |
AT5G45310
|
unknown |
-0.579 |
0.028 |
| 110 |
258017_at |
AT3G19370
|
unknown |
-0.575 |
-0.035 |
| 111 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
-0.572 |
-0.001 |
| 112 |
247037_at |
AT5G67070
|
RALFL34, ralf-like 34 |
-0.562 |
-0.092 |
| 113 |
265869_at |
AT2G01760
|
ARR14, response regulator 14, RR14, response regulator 14 |
-0.562 |
0.015 |
| 114 |
249118_at |
AT5G43870
|
[FUNCTIONS IN: phosphoinositide binding] |
-0.554 |
0.015 |
| 115 |
263902_at |
AT2G36230
|
APG10, ALBINO AND PALE GREEN 10, HISN3 |
-0.542 |
-0.098 |
| 116 |
251788_at |
AT3G55420
|
unknown |
-0.536 |
-0.142 |
| 117 |
265884_at |
AT2G42320
|
[nucleolar protein gar2-related] |
-0.535 |
-0.059 |
| 118 |
258196_at |
AT3G13980
|
unknown |
-0.534 |
-0.076 |
| 119 |
247069_at |
AT5G66920
|
sks17, SKU5 similar 17 |
-0.531 |
-0.068 |
| 120 |
262181_at |
AT1G78060
|
[Glycosyl hydrolase family protein] |
-0.527 |
-0.006 |
| 121 |
265938_at |
AT2G19620
|
NDL3, N-MYC downregulated-like 3 |
-0.526 |
0.020 |
| 122 |
260922_at |
AT1G21560
|
unknown |
-0.519 |
-0.040 |
| 123 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
-0.504 |
-0.045 |
| 124 |
255824_at |
AT2G40530
|
unknown |
-0.504 |
0.042 |
| 125 |
259430_at |
AT1G01610
|
ATGPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4, GPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4 |
-0.502 |
0.194 |
| 126 |
252184_at |
AT3G50660
|
AtDWF4, CLM, CLOMAZONE-RESISTANT, CYP90B1, CYTOCHROME P450 90B1, DWF4, DWARF 4, PSC1, PARTIALLY SUPPRESSING COI1 INSENSITIVITY TO JA 1, SAV1, SHADE AVOIDANCE 1, SNP2, SUPPRESSOR OF NPH4 2 |
-0.501 |
-0.230 |
| 127 |
267344_at |
AT2G44230
|
unknown |
-0.499 |
0.011 |
| 128 |
267310_at |
AT2G34680
|
AIR9, AUXIN-INDUCED IN ROOT CULTURES 9 |
-0.485 |
-0.022 |
| 129 |
248273_at |
AT5G53500
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.482 |
-0.003 |
| 130 |
249073_at |
AT5G44020
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.480 |
-0.037 |
| 131 |
258676_at |
AT3G08600
|
unknown |
-0.477 |
0.021 |
| 132 |
255488_at |
AT4G02630
|
[Protein kinase superfamily protein] |
-0.474 |
0.064 |
| 133 |
264371_at |
AT1G12090
|
ELP, extensin-like protein |
-0.468 |
-0.055 |
| 134 |
267040_at |
AT2G34300
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.468 |
-0.010 |
| 135 |
257236_at |
AT3G15095
|
HCF243, high chlorophyll fluorescence 243 |
-0.460 |
-0.008 |
| 136 |
259692_at |
AT1G63080
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.459 |
0.024 |
| 137 |
250975_at |
AT5G03050
|
unknown |
-0.454 |
-0.051 |
| 138 |
258432_at |
AT3G16570
|
ATRALF23, ARABIDOPSIS RAPID ALKALINIZATION FACTOR 23, RALF23, rapid alkalinization factor 23 |
-0.453 |
0.048 |
| 139 |
265844_at |
AT2G35620
|
FEI2, FEI 2 |
-0.453 |
-0.084 |
| 140 |
255948_at |
AT1G22060
|
[LOCATED IN: vacuole] |
-0.450 |
-0.010 |
| 141 |
256441_at |
AT3G10940
|
LSF2, LIKE SEX4 2 |
-0.444 |
0.040 |
| 142 |
261451_at |
AT1G21060
|
unknown |
-0.441 |
0.096 |
| 143 |
258368_at |
AT3G14240
|
[Subtilase family protein] |
-0.438 |
-0.042 |
| 144 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.433 |
-0.033 |
| 145 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
-0.429 |
-0.148 |
| 146 |
254193_at |
AT4G23870
|
unknown |
-0.425 |
0.089 |
| 147 |
247486_at |
AT5G62140
|
unknown |
-0.424 |
0.005 |
| 148 |
250102_at |
AT5G16590
|
LRR1, Leucine rich repeat protein 1 |
-0.424 |
-0.029 |
| 149 |
253903_at |
AT4G27180
|
ATK2, kinesin 2, KATB, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA B |
-0.423 |
-0.054 |
| 150 |
251676_at |
AT3G57320
|
unknown |
-0.418 |
0.020 |
| 151 |
247041_at |
AT5G67180
|
TOE3, target of early activation tagged (EAT) 3 |
-0.417 |
0.013 |
| 152 |
257516_at |
AT1G69040
|
ACR4, ACT domain repeat 4 |
-0.411 |
0.047 |
| 153 |
264360_at |
AT1G03310
|
AtBE2, ATISA2, ARABIDOPSIS THALIANA ISOAMYLASE 2, BE2, BRANCHING ENZYME 2, DBE1, debranching enzyme 1, ISA2 |
-0.410 |
-0.109 |
| 154 |
262312_at |
AT1G70830
|
MLP28, MLP-like protein 28 |
-0.410 |
-0.065 |
| 155 |
250245_at |
AT5G13690
|
CYL1, CYCLOPS 1, NAGLU, N-ACETYL-GLUCOSAMINIDASE |
-0.408 |
-0.029 |