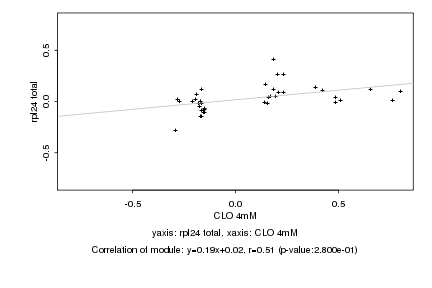
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
CLO 4mM |
Log2 signal ratio
rpl24 total |
| 1 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.800 |
0.100 |
| 2 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.763 |
0.010 |
| 3 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.654 |
0.118 |
| 4 |
246373_at |
AT1G51860
|
[Leucine-rich repeat protein kinase family protein] |
0.509 |
0.012 |
| 5 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
0.486 |
-0.003 |
| 6 |
259850_at |
AT1G72240
|
unknown |
0.482 |
0.042 |
| 7 |
265214_at |
AT1G05000
|
AtPFA-DSP1, PFA-DSP1, plant and fungi atypical dual-specificity phosphatase 1 |
0.419 |
0.113 |
| 8 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.386 |
0.146 |
| 9 |
255360_at |
AT4G03960
|
AtPFA-DSP4, PFA-DSP4, plant and fungi atypical dual-specificity phosphatase 4 |
0.231 |
0.090 |
| 10 |
252625_at |
AT3G44750
|
ATHD2A, HD2A, HISTONE DEACETYLASE 2A, HDA3, histone deacetylase 3, HDT1 |
0.231 |
0.272 |
| 11 |
248286_at |
AT5G52870
|
MAKR5, MEMBRANE-ASSOCIATED KINASE REGULATOR 5 |
0.205 |
0.094 |
| 12 |
265730_at |
AT2G32220
|
[Ribosomal L27e protein family] |
0.203 |
0.271 |
| 13 |
249306_at |
AT5G41400
|
[RING/U-box superfamily protein] |
0.194 |
0.055 |
| 14 |
254430_at |
AT4G20820
|
[FAD-binding Berberine family protein] |
0.183 |
0.412 |
| 15 |
250418_at |
AT5G11240
|
[transducin family protein / WD-40 repeat family protein] |
0.183 |
0.125 |
| 16 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.170 |
0.054 |
| 17 |
250415_at |
AT5G11210
|
ATGLR2.5, ARABIDOPSIS THALIANA GLU, GLR2.5, glutamate receptor 2.5 |
0.158 |
0.043 |
| 18 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.154 |
-0.010 |
| 19 |
247491_at |
AT5G61880
|
[Protein Transporter, Pam16] |
0.145 |
0.171 |
| 20 |
249068_at |
AT5G43980
|
PDLP1, plasmodesmata-located protein 1, PDLP1A, PLASMODESMATA-LOCATED PROTEIN 1A |
0.141 |
-0.001 |
| 21 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.293 |
-0.273 |
| 22 |
248344_at |
AT5G52280
|
[Myosin heavy chain-related protein] |
-0.286 |
0.029 |
| 23 |
247397_at |
AT5G62940
|
DOF5.6, DNA BINDING WITH ONE FINGER 5.6, HCA2, HIGH CAMBIAL ACTIVITY2 |
-0.272 |
0.005 |
| 24 |
249118_at |
AT5G43870
|
[FUNCTIONS IN: phosphoinositide binding] |
-0.213 |
0.006 |
| 25 |
265844_at |
AT2G35620
|
FEI2, FEI 2 |
-0.196 |
0.026 |
| 26 |
262785_at |
AT1G10750
|
unknown |
-0.194 |
0.069 |
| 27 |
262700_at |
AT1G76020
|
[Thioredoxin superfamily protein] |
-0.183 |
-0.011 |
| 28 |
258213_at |
AT3G17950
|
unknown |
-0.175 |
-0.046 |
| 29 |
267193_at |
AT2G30900
|
TBL43, TRICHOME BIREFRINGENCE-LIKE 43 |
-0.172 |
-0.144 |
| 30 |
261697_at |
AT1G32610
|
[hydroxyproline-rich glycoprotein family protein] |
-0.171 |
0.009 |
| 31 |
253581_at |
AT4G30660
|
[Low temperature and salt responsive protein family] |
-0.167 |
0.122 |
| 32 |
256323_at |
AT1G54920
|
unknown |
-0.167 |
-0.011 |
| 33 |
265253_at |
AT2G02020
|
AtNPF8.4, AtPTR4, NPF8.4, NRT1/ PTR family 8.4, PTR4, peptide transporter 4 |
-0.166 |
-0.085 |
| 34 |
263629_at |
AT2G04850
|
[Auxin-responsive family protein] |
-0.165 |
-0.138 |
| 35 |
267244_at |
AT2G02650
|
[Ribonuclease H-like superfamily protein] |
-0.160 |
-0.106 |
| 36 |
266216_at |
AT2G28810
|
[Dof-type zinc finger DNA-binding family protein] |
-0.157 |
-0.079 |
| 37 |
245386_at |
AT4G14010
|
RALFL32, ralf-like 32 |
-0.151 |
-0.107 |
| 38 |
247856_at |
AT5G58300
|
[Leucine-rich repeat protein kinase family protein] |
-0.151 |
-0.074 |
| 39 |
259704_at |
AT1G77680
|
[Ribonuclease II/R family protein] |
-0.151 |
-0.064 |