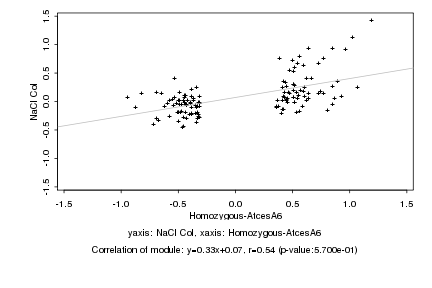
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Homozygous-AtcesA6 |
Log2 signal ratio
NaCl Col |
| 1 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
1.185 |
1.441 |
| 2 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
1.065 |
0.254 |
| 3 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.022 |
1.132 |
| 4 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.960 |
0.915 |
| 5 |
249117_at |
AT5G43840
|
AT-HSFA6A, heat shock transcription factor A6A, HSFA6A, heat shock transcription factor A6A |
0.922 |
0.099 |
| 6 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.888 |
0.362 |
| 7 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.861 |
0.064 |
| 8 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
0.847 |
0.266 |
| 9 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.847 |
0.939 |
| 10 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
0.844 |
-0.041 |
| 11 |
259866_at |
AT1G76640
|
CML39, CALMODULIN LIKE 39 |
0.799 |
-0.150 |
| 12 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.769 |
0.148 |
| 13 |
263881_at |
AT2G21820
|
unknown |
0.769 |
0.760 |
| 14 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.742 |
0.181 |
| 15 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.724 |
0.143 |
| 16 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
0.719 |
0.684 |
| 17 |
264580_at |
AT1G05340
|
unknown |
0.663 |
0.417 |
| 18 |
255527_at |
AT4G02360
|
unknown |
0.638 |
0.070 |
| 19 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
0.633 |
0.939 |
| 20 |
255900_at |
AT1G17830
|
unknown |
0.631 |
0.148 |
| 21 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
0.614 |
0.414 |
| 22 |
261922_at |
AT1G65890
|
AAE12, acyl activating enzyme 12 |
0.614 |
0.033 |
| 23 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
0.603 |
0.251 |
| 24 |
259054_at |
AT3G03480
|
CHAT, acetyl CoA:(Z)-3-hexen-1-ol acetyltransferase |
0.598 |
0.094 |
| 25 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.595 |
0.642 |
| 26 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
0.591 |
0.191 |
| 27 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.580 |
-0.087 |
| 28 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.566 |
0.207 |
| 29 |
256875_at |
AT3G26330
|
CYP71B37, cytochrome P450, family 71, subfamily B, polypeptide 37 |
0.556 |
-0.171 |
| 30 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.555 |
0.802 |
| 31 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
0.548 |
0.116 |
| 32 |
250158_at |
AT5G15190
|
unknown |
0.541 |
0.684 |
| 33 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
0.538 |
0.054 |
| 34 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.532 |
0.169 |
| 35 |
257595_at |
AT3G24750
|
unknown |
0.528 |
-0.190 |
| 36 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.514 |
-0.017 |
| 37 |
250826_at |
AT5G05220
|
unknown |
0.513 |
0.614 |
| 38 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.511 |
-0.004 |
| 39 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.509 |
0.295 |
| 40 |
257450_at |
AT1G10530
|
unknown |
0.504 |
0.304 |
| 41 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
0.504 |
0.087 |
| 42 |
253293_at |
AT4G33905
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.504 |
0.537 |
| 43 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
0.503 |
0.314 |
| 44 |
259439_at |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
0.503 |
0.207 |
| 45 |
250103_at |
AT5G16600
|
AtMYB43, myb domain protein 43, MYB43, myb domain protein 43 |
0.503 |
0.075 |
| 46 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.495 |
0.727 |
| 47 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.473 |
0.152 |
| 48 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
0.470 |
0.546 |
| 49 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.461 |
0.160 |
| 50 |
262236_at |
AT1G48330
|
unknown |
0.453 |
0.068 |
| 51 |
251062_at |
AT5G01840
|
ATOFP1, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 1, OFP1, ovate family protein 1 |
0.453 |
-0.005 |
| 52 |
257460_at |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
0.441 |
0.279 |
| 53 |
247360_at |
AT5G63450
|
CYP94B1, cytochrome P450, family 94, subfamily B, polypeptide 1 |
0.439 |
0.034 |
| 54 |
256603_at |
AT3G28270
|
unknown |
0.439 |
0.052 |
| 55 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.430 |
0.350 |
| 56 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
0.425 |
0.162 |
| 57 |
260948_at |
AT1G06100
|
[Fatty acid desaturase family protein] |
0.423 |
0.078 |
| 58 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.421 |
0.078 |
| 59 |
252488_at |
AT3G46700
|
[UDP-Glycosyltransferase superfamily protein] |
0.415 |
-0.138 |
| 60 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.414 |
0.358 |
| 61 |
247047_at |
AT5G66650
|
unknown |
0.412 |
0.097 |
| 62 |
252321_at |
AT3G48510
|
unknown |
0.411 |
0.254 |
| 63 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
0.409 |
-0.127 |
| 64 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
0.409 |
0.020 |
| 65 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
0.400 |
-0.207 |
| 66 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.380 |
0.757 |
| 67 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
0.374 |
-0.074 |
| 68 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
0.365 |
0.028 |
| 69 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
0.354 |
-0.086 |
| 70 |
253172_at |
AT4G35060
|
HIPP25, heavy metal associated isoprenylated plant protein 25 |
0.353 |
-0.097 |
| 71 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.953 |
0.082 |
| 72 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
-0.881 |
-0.104 |
| 73 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
-0.826 |
0.155 |
| 74 |
258468_at |
AT3G06070
|
unknown |
-0.718 |
-0.392 |
| 75 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-0.697 |
-0.292 |
| 76 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
-0.696 |
0.172 |
| 77 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
-0.681 |
-0.333 |
| 78 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
-0.654 |
0.149 |
| 79 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
-0.628 |
-0.079 |
| 80 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
-0.599 |
-0.032 |
| 81 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
-0.582 |
0.019 |
| 82 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.581 |
-0.250 |
| 83 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
-0.557 |
0.038 |
| 84 |
254032_at |
AT4G25940
|
[ENTH/ANTH/VHS superfamily protein] |
-0.546 |
-0.061 |
| 85 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
-0.542 |
0.408 |
| 86 |
252346_at |
AT3G48650
|
[pseudogene] |
-0.537 |
0.079 |
| 87 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.523 |
-0.034 |
| 88 |
246993_at |
AT5G67450
|
AZF1, zinc-finger protein 1, ZF1, zinc-finger protein 1 |
-0.508 |
-0.184 |
| 89 |
258653_at |
AT3G09870
|
SAUR48, SMALL AUXIN UPREGULATED RNA 48 |
-0.507 |
0.025 |
| 90 |
247189_at |
AT5G65390
|
AGP7, arabinogalactan protein 7 |
-0.504 |
-0.338 |
| 91 |
257076_at |
AT3G19680
|
unknown |
-0.501 |
-0.176 |
| 92 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
-0.500 |
-0.187 |
| 93 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.497 |
0.035 |
| 94 |
250446_at |
AT5G10770
|
[Eukaryotic aspartyl protease family protein] |
-0.492 |
0.163 |
| 95 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.492 |
-0.040 |
| 96 |
264232_at |
AT1G67470
|
[Protein kinase superfamily protein] |
-0.481 |
-0.180 |
| 97 |
254948_at |
AT4G11000
|
[Ankyrin repeat family protein] |
-0.479 |
-0.166 |
| 98 |
256593_at |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.478 |
-0.049 |
| 99 |
267034_at |
AT2G38310
|
PYL4, PYR1-like 4, RCAR10, regulatory components of ABA receptor 10 |
-0.472 |
-0.444 |
| 100 |
254266_at |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
-0.462 |
0.034 |
| 101 |
250582_at |
AT5G07580
|
[Integrase-type DNA-binding superfamily protein] |
-0.459 |
-0.435 |
| 102 |
247474_at |
AT5G62280
|
unknown |
-0.455 |
-0.270 |
| 103 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
-0.451 |
-0.011 |
| 104 |
248870_at |
AT5G46710
|
[PLATZ transcription factor family protein] |
-0.451 |
0.113 |
| 105 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
-0.446 |
0.074 |
| 106 |
260975_at |
AT1G53430
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.444 |
-0.188 |
| 107 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.442 |
-0.041 |
| 108 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-0.438 |
0.117 |
| 109 |
260427_at |
AT1G72430
|
SAUR78, SMALL AUXIN UPREGULATED RNA 78 |
-0.436 |
-0.298 |
| 110 |
245840_at |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
-0.436 |
-0.036 |
| 111 |
257636_at |
AT3G26200
|
CYP71B22, cytochrome P450, family 71, subfamily B, polypeptide 22 |
-0.433 |
-0.057 |
| 112 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
-0.427 |
0.029 |
| 113 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
-0.405 |
-0.221 |
| 114 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
-0.395 |
-0.030 |
| 115 |
260735_at |
AT1G17610
|
CHS1, CHILLING SENSITIVE 1 |
-0.395 |
-0.006 |
| 116 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
-0.391 |
0.216 |
| 117 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
-0.390 |
-0.194 |
| 118 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
-0.388 |
-0.103 |
| 119 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
-0.387 |
0.101 |
| 120 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
-0.386 |
-0.222 |
| 121 |
264590_at |
AT2G17710
|
unknown |
-0.372 |
0.018 |
| 122 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
-0.368 |
0.067 |
| 123 |
263565_at |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
-0.356 |
-0.205 |
| 124 |
266175_at |
AT2G02450
|
ANAC034, Arabidopsis NAC domain containing protein 34, ANAC035, NAC domain containing protein 35, AtLOV1, LOV1, LONG VEGETATIVE PHASE 1, NAC035, NAC domain containing protein 35 |
-0.355 |
-0.057 |
| 125 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-0.349 |
-0.358 |
| 126 |
261491_at |
AT1G14350
|
AtMYB124, myb domain protein 124, FLP, FOUR LIPS, MYB124 |
-0.346 |
-0.095 |
| 127 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
-0.342 |
0.247 |
| 128 |
264704_at |
AT1G70090
|
GATL9, GALACTURONOSYLTRANSFERASE-LIKE 9, LGT8, glucosyl transferase family 8 |
-0.340 |
-0.286 |
| 129 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
-0.339 |
-0.071 |
| 130 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.333 |
-0.185 |
| 131 |
252976_s_at |
AT4G38550
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
-0.333 |
-0.082 |
| 132 |
264624_at |
AT1G08930
|
ERD6, EARLY RESPONSE TO DEHYDRATION 6 |
-0.330 |
-0.002 |
| 133 |
248179_at |
AT5G54380
|
THE1, THESEUS1 |
-0.329 |
-0.253 |
| 134 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.326 |
-0.227 |
| 135 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
-0.321 |
-0.029 |
| 136 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
-0.318 |
-0.081 |
| 137 |
263379_at |
AT2G40140
|
ATSZF2, CZF1, SZF2, (SALT-INDUCIBLE ZINC FINGER 2, ZFAR1 |
-0.318 |
0.098 |
| 138 |
252944_at |
AT4G39320
|
[microtubule-associated protein-related] |
-0.317 |
-0.006 |
| 139 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
-0.316 |
-0.270 |