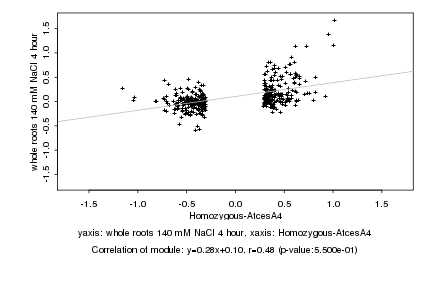
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Homozygous-AtcesA4 |
Log2 signal ratio
whole roots 140 mM NaCl 4 hour |
| 1 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
1.005 |
1.687 |
| 2 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.000 |
1.164 |
| 3 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.952 |
1.384 |
| 4 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.917 |
0.122 |
| 5 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
0.818 |
0.504 |
| 6 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.811 |
0.194 |
| 7 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
0.793 |
0.028 |
| 8 |
265359_at |
AT2G16720
|
ATMYB7, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 7, ATY49, MYB7, myb domain protein 7 |
0.754 |
0.182 |
| 9 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.732 |
0.167 |
| 10 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.726 |
1.144 |
| 11 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.710 |
0.421 |
| 12 |
255527_at |
AT4G02360
|
unknown |
0.698 |
0.148 |
| 13 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
0.653 |
0.365 |
| 14 |
264580_at |
AT1G05340
|
unknown |
0.648 |
0.481 |
| 15 |
265024_at |
AT1G24600
|
unknown |
0.647 |
0.532 |
| 16 |
245444_at |
AT4G16740
|
ATTPS03, terpene synthase 03, TPS03, terpene synthase 03 |
0.639 |
0.025 |
| 17 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.625 |
0.195 |
| 18 |
263881_at |
AT2G21820
|
unknown |
0.624 |
0.540 |
| 19 |
259866_at |
AT1G76640
|
CML39, CALMODULIN LIKE 39 |
0.623 |
0.040 |
| 20 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.622 |
0.008 |
| 21 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.619 |
0.574 |
| 22 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.615 |
0.220 |
| 23 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
0.614 |
0.496 |
| 24 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.609 |
1.147 |
| 25 |
253293_at |
AT4G33905
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.607 |
0.578 |
| 26 |
254773_at |
AT4G13410
|
ATCSLA15, CSLA15, CELLULOSE SYNTHASE LIKE A15 |
0.607 |
-0.074 |
| 27 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.602 |
0.381 |
| 28 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.597 |
0.401 |
| 29 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.596 |
0.805 |
| 30 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
0.590 |
0.483 |
| 31 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
0.581 |
0.228 |
| 32 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.572 |
0.130 |
| 33 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
0.568 |
0.906 |
| 34 |
263544_at |
AT2G21590
|
APL4 |
0.566 |
0.077 |
| 35 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.560 |
0.767 |
| 36 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.559 |
0.566 |
| 37 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.557 |
0.560 |
| 38 |
249771_at |
AT5G24080
|
[Protein kinase superfamily protein] |
0.552 |
0.777 |
| 39 |
261922_at |
AT1G65890
|
AAE12, acyl activating enzyme 12 |
0.551 |
0.054 |
| 40 |
265030_at |
AT1G61610
|
[S-locus lectin protein kinase family protein] |
0.546 |
0.009 |
| 41 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.543 |
0.279 |
| 42 |
263486_at |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
0.540 |
0.202 |
| 43 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.538 |
0.030 |
| 44 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.537 |
0.062 |
| 45 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.531 |
0.079 |
| 46 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
0.530 |
0.039 |
| 47 |
262236_at |
AT1G48330
|
unknown |
0.525 |
0.136 |
| 48 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
0.523 |
0.019 |
| 49 |
256296_at |
AT1G69480
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.522 |
0.617 |
| 50 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
0.521 |
0.163 |
| 51 |
266108_at |
AT2G37900
|
[Major facilitator superfamily protein] |
0.512 |
-0.064 |
| 52 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.507 |
0.716 |
| 53 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
0.506 |
0.376 |
| 54 |
266661_at |
AT2G25820
|
ESE2, ethylene and salt inducible 2 |
0.506 |
-0.003 |
| 55 |
256603_at |
AT3G28270
|
unknown |
0.485 |
0.043 |
| 56 |
247360_at |
AT5G63450
|
CYP94B1, cytochrome P450, family 94, subfamily B, polypeptide 1 |
0.481 |
-0.042 |
| 57 |
260420_at |
AT1G69610
|
[FUNCTIONS IN: structural constituent of ribosome] |
0.478 |
0.154 |
| 58 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.477 |
0.519 |
| 59 |
263031_at |
AT1G24070
|
ATCSLA10, cellulose synthase-like A10, CSLA10, CELLULOSE SYNTHASE LIKE A10, CSLA10, cellulose synthase-like A10 |
0.473 |
-0.015 |
| 60 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
0.464 |
0.075 |
| 61 |
258652_at |
AT3G09910
|
ATRAB18C, ATRABC2B, RAB GTPase homolog C2B, RABC2b, RAB GTPase homolog C2B |
0.462 |
0.468 |
| 62 |
247511_at |
AT5G62040
|
BFT, brother of FT and TFL1 |
0.462 |
0.123 |
| 63 |
257951_at |
AT3G21700
|
ATSGP2, SGP2 |
0.462 |
0.151 |
| 64 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.461 |
0.315 |
| 65 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
0.459 |
0.515 |
| 66 |
249540_at |
AT5G38120
|
4CL8 |
0.457 |
0.019 |
| 67 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.456 |
-0.208 |
| 68 |
263005_at |
AT1G54540
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.453 |
0.153 |
| 69 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
0.453 |
-0.019 |
| 70 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.449 |
0.107 |
| 71 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.443 |
0.079 |
| 72 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.439 |
0.161 |
| 73 |
253799_at |
AT4G28140
|
[Integrase-type DNA-binding superfamily protein] |
0.438 |
0.177 |
| 74 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.437 |
0.340 |
| 75 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.436 |
0.682 |
| 76 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.432 |
0.160 |
| 77 |
250158_at |
AT5G15190
|
unknown |
0.426 |
0.449 |
| 78 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.423 |
0.032 |
| 79 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.417 |
0.151 |
| 80 |
257017_at |
AT3G19620
|
[Glycosyl hydrolase family protein] |
0.417 |
0.002 |
| 81 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
0.412 |
-0.151 |
| 82 |
253619_at |
AT4G30460
|
[glycine-rich protein] |
0.407 |
0.236 |
| 83 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
0.403 |
0.144 |
| 84 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.401 |
0.606 |
| 85 |
260005_at |
AT1G67920
|
unknown |
0.401 |
0.138 |
| 86 |
250483_at |
AT5G10300
|
AtHNL, ATMES5, ARABIDOPSIS THALIANA METHYL ESTERASE 5, HNL, HYDROXYNITRILE LYASE, MES5, methyl esterase 5 |
0.399 |
0.243 |
| 87 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.399 |
0.685 |
| 88 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.398 |
0.758 |
| 89 |
259618_at |
AT1G48000
|
AtMYB112, myb domain protein 112, MYB112, myb domain protein 112 |
0.398 |
0.543 |
| 90 |
266555_at |
AT2G46270
|
GBF3, G-box binding factor 3 |
0.396 |
0.099 |
| 91 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
0.395 |
0.164 |
| 92 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
0.394 |
-0.104 |
| 93 |
249541_at |
AT5G38130
|
[HXXXD-type acyl-transferase family protein] |
0.391 |
-0.013 |
| 94 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
0.391 |
0.091 |
| 95 |
261224_at |
AT1G20160
|
ATSBT5.2 |
0.389 |
-0.117 |
| 96 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
0.386 |
-0.130 |
| 97 |
261586_at |
AT1G01640
|
[BTB/POZ domain-containing protein] |
0.385 |
-0.095 |
| 98 |
262061_at |
AT1G80110
|
ATPP2-B11, phloem protein 2-B11, PP2-B11, phloem protein 2-B11 |
0.384 |
0.445 |
| 99 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
0.381 |
-0.044 |
| 100 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
0.379 |
0.062 |
| 101 |
261247_at |
AT1G20070
|
unknown |
0.376 |
0.259 |
| 102 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.375 |
0.670 |
| 103 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
0.375 |
0.049 |
| 104 |
252365_at |
AT3G48350
|
CEP3, cysteine endopeptidase 3 |
0.374 |
-0.075 |
| 105 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
0.374 |
0.027 |
| 106 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.373 |
-0.063 |
| 107 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.372 |
0.252 |
| 108 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.371 |
0.338 |
| 109 |
253842_at |
AT4G27860
|
MEB1, MEMBRANE OF ER BODY 1 |
0.371 |
-0.211 |
| 110 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
0.371 |
0.114 |
| 111 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
0.369 |
0.051 |
| 112 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.369 |
0.222 |
| 113 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.368 |
0.439 |
| 114 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.366 |
0.289 |
| 115 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.363 |
0.024 |
| 116 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
0.363 |
0.204 |
| 117 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.362 |
0.157 |
| 118 |
245699_at |
AT5G04250
|
[Cysteine proteinases superfamily protein] |
0.362 |
0.264 |
| 119 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.361 |
0.502 |
| 120 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
0.361 |
-0.046 |
| 121 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.361 |
-0.107 |
| 122 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
0.361 |
0.301 |
| 123 |
257450_at |
AT1G10530
|
unknown |
0.361 |
0.252 |
| 124 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
0.359 |
0.199 |
| 125 |
254996_at |
AT4G10390
|
[Protein kinase superfamily protein] |
0.359 |
-0.118 |
| 126 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.358 |
0.819 |
| 127 |
246582_at |
AT1G31750
|
[proline-rich family protein] |
0.354 |
0.436 |
| 128 |
245831_at |
AT1G48840
|
unknown |
0.353 |
-0.061 |
| 129 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
0.351 |
0.232 |
| 130 |
246125_at |
AT5G19875
|
unknown |
0.350 |
0.292 |
| 131 |
266321_at |
AT2G46660
|
CYP78A6, cytochrome P450, family 78, subfamily A, polypeptide 6, EOD3, enhancer of da1-1 |
0.349 |
0.012 |
| 132 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
0.348 |
-0.005 |
| 133 |
258878_at |
AT3G03170
|
unknown |
0.347 |
0.327 |
| 134 |
259149_at |
AT3G10340
|
PAL4, phenylalanine ammonia-lyase 4 |
0.344 |
0.144 |
| 135 |
260361_at |
AT1G69360
|
unknown |
0.343 |
0.109 |
| 136 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.342 |
0.047 |
| 137 |
247199_at |
AT5G65210
|
TGA1, TGACG sequence-specific binding protein 1 |
0.342 |
0.008 |
| 138 |
257035_at |
AT3G19270
|
CYP707A4, cytochrome P450, family 707, subfamily A, polypeptide 4 |
0.341 |
0.088 |
| 139 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.341 |
0.138 |
| 140 |
262211_at |
AT1G74930
|
ORA47 |
0.338 |
0.074 |
| 141 |
259439_at |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
0.338 |
-0.003 |
| 142 |
265059_at |
AT1G52080
|
AR791 |
0.338 |
0.135 |
| 143 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
0.331 |
0.821 |
| 144 |
247030_at |
AT5G67210
|
IRX15-L, IRX15-LIKE |
0.331 |
0.006 |
| 145 |
248896_at |
AT5G46350
|
ATWRKY8, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 8, WRKY8, WRKY DNA-binding protein 8 |
0.331 |
0.109 |
| 146 |
251591_at |
AT3G57680
|
[Peptidase S41 family protein] |
0.330 |
0.053 |
| 147 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.330 |
0.028 |
| 148 |
245445_at |
AT4G16750
|
[Integrase-type DNA-binding superfamily protein] |
0.329 |
0.174 |
| 149 |
250127_at |
AT5G16380
|
unknown |
0.327 |
0.142 |
| 150 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
0.327 |
0.142 |
| 151 |
260264_at |
AT1G68500
|
unknown |
0.326 |
0.181 |
| 152 |
265452_at |
AT2G46510
|
AIB, ABA-inducible BHLH-type transcription factor, ATAIB, ABA-inducible BHLH-type transcription factor, JAM1, JA-associated MYC2-like 1 |
0.325 |
0.115 |
| 153 |
265334_at |
AT2G18370
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.325 |
0.100 |
| 154 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.325 |
0.187 |
| 155 |
256937_at |
AT3G22620
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.325 |
0.137 |
| 156 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
0.324 |
0.254 |
| 157 |
259043_at |
AT3G03440
|
[ARM repeat superfamily protein] |
0.322 |
0.221 |
| 158 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.321 |
0.100 |
| 159 |
256114_at |
AT1G16850
|
unknown |
0.320 |
0.623 |
| 160 |
267509_at |
AT2G45660
|
AGL20, AGAMOUS-like 20, ATSOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1, SOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1 |
0.320 |
0.016 |
| 161 |
252309_at |
AT3G49340
|
[Cysteine proteinases superfamily protein] |
0.318 |
-0.014 |
| 162 |
256021_at |
AT1G58270
|
ZW9 |
0.318 |
0.081 |
| 163 |
264252_at |
AT1G09180
|
ATSAR1, SECRETION-ASSOCIATED RAS 1, ATSARA1A, secretion-associated RAS super family 1, SARA1A, secretion-associated RAS super family 1 |
0.318 |
0.315 |
| 164 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.317 |
0.122 |
| 165 |
251291_at |
AT3G61900
|
SAUR33, SMALL AUXIN UPREGULATED RNA 33 |
0.316 |
0.048 |
| 166 |
247312_at |
AT5G63970
|
RGLG3, RING DOMAIN LIGASE 3 |
0.316 |
-0.043 |
| 167 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.316 |
0.279 |
| 168 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
0.315 |
-0.028 |
| 169 |
252611_at |
AT3G45130
|
LAS1, lanosterol synthase 1 |
0.314 |
-0.088 |
| 170 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.314 |
-0.029 |
| 171 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.313 |
0.157 |
| 172 |
261748_at |
AT1G76070
|
unknown |
0.311 |
-0.040 |
| 173 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
0.310 |
0.038 |
| 174 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.308 |
0.728 |
| 175 |
253407_at |
AT4G32920
|
[glycine-rich protein] |
0.307 |
0.122 |
| 176 |
264887_at |
AT1G23120
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.307 |
0.228 |
| 177 |
248505_at |
AT5G50360
|
unknown |
0.306 |
0.560 |
| 178 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
0.304 |
0.253 |
| 179 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
0.304 |
-0.044 |
| 180 |
251725_at |
AT3G56260
|
unknown |
0.304 |
0.065 |
| 181 |
250589_at |
AT5G07700
|
AtMYB76, myb domain protein 76, MYB76, myb domain protein 76 |
0.304 |
0.003 |
| 182 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.303 |
0.099 |
| 183 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
0.303 |
0.040 |
| 184 |
264314_at |
AT1G70420
|
unknown |
0.303 |
-0.056 |
| 185 |
251768_at |
AT3G55940
|
[Phosphoinositide-specific phospholipase C family protein] |
0.302 |
-0.050 |
| 186 |
250955_at |
AT5G03190
|
CPUORF47, conserved peptide upstream open reading frame 47 |
0.302 |
0.141 |
| 187 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.302 |
0.390 |
| 188 |
261803_at |
AT1G30500
|
NF-YA7, nuclear factor Y, subunit A7 |
0.301 |
0.136 |
| 189 |
261758_at |
AT1G08250
|
ADT6, arogenate dehydratase 6, AtADT6, Arabidopsis thaliana arogenate dehydratase 6 |
0.297 |
0.133 |
| 190 |
258374_at |
AT3G14360
|
[alpha/beta-Hydrolases superfamily protein] |
0.297 |
0.072 |
| 191 |
258209_at |
AT3G14060
|
unknown |
0.297 |
0.045 |
| 192 |
250211_at |
AT5G13880
|
unknown |
0.296 |
0.331 |
| 193 |
248959_at |
AT5G45630
|
unknown |
0.296 |
0.050 |
| 194 |
259802_at |
AT1G72260
|
THI2.1, thionin 2.1, THI2.1.1 |
0.295 |
-0.073 |
| 195 |
252321_at |
AT3G48510
|
unknown |
0.295 |
0.559 |
| 196 |
257460_at |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
0.294 |
-0.026 |
| 197 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
0.291 |
-0.042 |
| 198 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.291 |
0.434 |
| 199 |
261266_at |
AT1G26770
|
AT-EXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXPA10, expansin A10, ATHEXP ALPHA 1.1, ARABIDOPSIS THALIANA EXPANSIN ALPHA 1.1, EXP10, EXPANSIN 10, EXPA10, expansin A10 |
0.291 |
0.375 |
| 200 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
0.290 |
0.080 |
| 201 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
0.290 |
0.249 |
| 202 |
251200_at |
AT3G63010
|
ATGID1B, GID1B, GA INSENSITIVE DWARF1B |
0.288 |
0.095 |
| 203 |
263325_at |
AT2G04240
|
XERICO |
0.288 |
-0.020 |
| 204 |
264479_at |
AT1G77280
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
0.287 |
-0.096 |
| 205 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-1.162 |
0.288 |
| 206 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
-1.045 |
0.031 |
| 207 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
-1.037 |
0.087 |
| 208 |
261658_at |
AT1G50040
|
unknown |
-0.824 |
0.005 |
| 209 |
250936_at |
AT5G03120
|
unknown |
-0.817 |
0.002 |
| 210 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.745 |
0.071 |
| 211 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
-0.737 |
0.049 |
| 212 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
-0.737 |
-0.181 |
| 213 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
-0.727 |
0.434 |
| 214 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
-0.716 |
0.113 |
| 215 |
254770_at |
AT4G13340
|
LRX3, leucine-rich repeat/extensin 3 |
-0.710 |
0.009 |
| 216 |
246200_at |
AT4G37240
|
unknown |
-0.709 |
-0.190 |
| 217 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
-0.697 |
-0.029 |
| 218 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-0.695 |
0.351 |
| 219 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
-0.678 |
-0.078 |
| 220 |
261984_at |
AT1G33760
|
[Integrase-type DNA-binding superfamily protein] |
-0.636 |
0.006 |
| 221 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
-0.631 |
-0.160 |
| 222 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
-0.630 |
-0.237 |
| 223 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.624 |
0.141 |
| 224 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
-0.623 |
-0.155 |
| 225 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.622 |
-0.059 |
| 226 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
-0.619 |
0.266 |
| 227 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-0.609 |
0.128 |
| 228 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
-0.606 |
-0.033 |
| 229 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
-0.606 |
0.185 |
| 230 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
-0.599 |
-0.132 |
| 231 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.596 |
-0.095 |
| 232 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
-0.596 |
-0.145 |
| 233 |
258653_at |
AT3G09870
|
SAUR48, SMALL AUXIN UPREGULATED RNA 48 |
-0.590 |
-0.065 |
| 234 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
-0.588 |
-0.136 |
| 235 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.584 |
0.037 |
| 236 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
-0.584 |
-0.019 |
| 237 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.583 |
-0.459 |
| 238 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
-0.571 |
0.094 |
| 239 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
-0.570 |
0.276 |
| 240 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
-0.567 |
0.065 |
| 241 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
-0.564 |
0.037 |
| 242 |
259729_at |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
-0.558 |
0.025 |
| 243 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.557 |
-0.309 |
| 244 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.557 |
0.067 |
| 245 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
-0.548 |
-0.196 |
| 246 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
-0.548 |
-0.040 |
| 247 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.545 |
-0.139 |
| 248 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.544 |
-0.175 |
| 249 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.544 |
0.200 |
| 250 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-0.538 |
0.045 |
| 251 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
-0.535 |
0.064 |
| 252 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.533 |
-0.081 |
| 253 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.525 |
-0.052 |
| 254 |
267393_at |
AT2G44500
|
[O-fucosyltransferase family protein] |
-0.524 |
-0.007 |
| 255 |
267034_at |
AT2G38310
|
PYL4, PYR1-like 4, RCAR10, regulatory components of ABA receptor 10 |
-0.515 |
-0.255 |
| 256 |
261926_at |
AT1G22530
|
PATL2, PATELLIN 2 |
-0.515 |
-0.232 |
| 257 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.514 |
-0.034 |
| 258 |
264770_at |
AT1G23030
|
[ARM repeat superfamily protein] |
-0.513 |
-0.126 |
| 259 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-0.512 |
-0.262 |
| 260 |
260975_at |
AT1G53430
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.511 |
-0.178 |
| 261 |
266956_at |
AT2G34510
|
unknown |
-0.507 |
0.034 |
| 262 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
-0.503 |
-0.235 |
| 263 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
-0.503 |
0.089 |
| 264 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.500 |
0.019 |
| 265 |
264704_at |
AT1G70090
|
GATL9, GALACTURONOSYLTRANSFERASE-LIKE 9, LGT8, glucosyl transferase family 8 |
-0.499 |
-0.006 |
| 266 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
-0.496 |
0.288 |
| 267 |
257076_at |
AT3G19680
|
unknown |
-0.493 |
-0.038 |
| 268 |
263421_at |
AT2G17230
|
EXL5, EXORDIUM like 5 |
-0.493 |
0.042 |
| 269 |
256633_at |
AT3G28340
|
GATL10, galacturonosyltransferase-like 10, GolS8, galactinol synthase 8 |
-0.493 |
0.123 |
| 270 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
-0.488 |
0.455 |
| 271 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
-0.488 |
0.065 |
| 272 |
259325_at |
AT3G05320
|
[O-fucosyltransferase family protein] |
-0.487 |
0.253 |
| 273 |
258468_at |
AT3G06070
|
unknown |
-0.486 |
-0.003 |
| 274 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
-0.486 |
-0.048 |
| 275 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
-0.485 |
-0.222 |
| 276 |
249073_at |
AT5G44020
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.485 |
-0.114 |
| 277 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
-0.485 |
-0.260 |
| 278 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
-0.480 |
0.027 |
| 279 |
260296_at |
AT1G63750
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.475 |
0.016 |
| 280 |
258225_at |
AT3G15630
|
unknown |
-0.467 |
-0.268 |
| 281 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
-0.466 |
-0.255 |
| 282 |
260656_at |
AT1G19380
|
unknown |
-0.465 |
0.071 |
| 283 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
-0.463 |
-0.033 |
| 284 |
263182_at |
AT1G05575
|
unknown |
-0.461 |
-0.060 |
| 285 |
264521_at |
AT1G10020
|
unknown |
-0.459 |
-0.085 |
| 286 |
252053_at |
AT3G52400
|
ATSYP122, SYP122, syntaxin of plants 122 |
-0.457 |
0.030 |
| 287 |
245176_at |
AT2G47440
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.456 |
-0.037 |
| 288 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
-0.456 |
0.097 |
| 289 |
247189_at |
AT5G65390
|
AGP7, arabinogalactan protein 7 |
-0.454 |
0.025 |
| 290 |
260427_at |
AT1G72430
|
SAUR78, SMALL AUXIN UPREGULATED RNA 78 |
-0.453 |
-0.194 |
| 291 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
-0.450 |
0.204 |
| 292 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
-0.448 |
0.026 |
| 293 |
253811_at |
AT4G28190
|
ULT, ULTRAPETALA, ULT1, ULTRAPETALA1 |
-0.447 |
-0.064 |
| 294 |
265732_at |
AT2G01300
|
unknown |
-0.447 |
0.212 |
| 295 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
-0.447 |
0.065 |
| 296 |
264624_at |
AT1G08930
|
ERD6, EARLY RESPONSE TO DEHYDRATION 6 |
-0.443 |
-0.039 |
| 297 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
-0.439 |
-0.131 |
| 298 |
250777_at |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
-0.438 |
-0.215 |
| 299 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.437 |
-0.021 |
| 300 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.426 |
0.010 |
| 301 |
267209_at |
AT2G30930
|
unknown |
-0.422 |
-0.107 |
| 302 |
252346_at |
AT3G48650
|
[pseudogene] |
-0.422 |
0.036 |
| 303 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.422 |
-0.105 |
| 304 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.415 |
-0.582 |
| 305 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
-0.414 |
-0.005 |
| 306 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.414 |
0.062 |
| 307 |
264232_at |
AT1G67470
|
[Protein kinase superfamily protein] |
-0.414 |
-0.044 |
| 308 |
249872_at |
AT5G23130
|
[Peptidoglycan-binding LysM domain-containing protein] |
-0.412 |
0.048 |
| 309 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.412 |
0.294 |
| 310 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
-0.412 |
0.070 |
| 311 |
266946_at |
AT2G18890
|
[Protein kinase superfamily protein] |
-0.411 |
0.147 |
| 312 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.410 |
-0.061 |
| 313 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
-0.409 |
-0.131 |
| 314 |
248179_at |
AT5G54380
|
THE1, THESEUS1 |
-0.406 |
-0.036 |
| 315 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
-0.405 |
-0.258 |
| 316 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.404 |
0.148 |
| 317 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
-0.401 |
0.031 |
| 318 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
-0.399 |
0.011 |
| 319 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.399 |
-0.026 |
| 320 |
256848_at |
AT3G27960
|
KLCR2, kinesin light chain-related 2 |
-0.398 |
0.016 |
| 321 |
257140_at |
AT3G28910
|
ATMYB30, MYB30, myb domain protein 30 |
-0.395 |
0.016 |
| 322 |
247878_at |
AT5G57760
|
unknown |
-0.393 |
-0.054 |
| 323 |
253514_at |
AT4G31805
|
POLAR, POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION |
-0.392 |
-0.013 |
| 324 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.392 |
-0.511 |
| 325 |
254041_at |
AT4G25830
|
[Uncharacterised protein family (UPF0497)] |
-0.388 |
-0.138 |
| 326 |
245084_at |
AT2G23290
|
AtMYB70, myb domain protein 70, MYB70, myb domain protein 70 |
-0.387 |
-0.048 |
| 327 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
-0.387 |
-0.020 |
| 328 |
264195_at |
AT1G22690
|
[Gibberellin-regulated family protein] |
-0.386 |
0.039 |
| 329 |
259329_at |
AT3G16360
|
AHP4, HPT phosphotransmitter 4 |
-0.385 |
0.411 |
| 330 |
255549_at |
AT4G01950
|
ATGPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3, GPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3 |
-0.382 |
0.109 |
| 331 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
-0.381 |
-0.026 |
| 332 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-0.380 |
-0.144 |
| 333 |
255538_at |
AT4G01680
|
AtMYB55, myb domain protein 55, MYB55, myb domain protein 55 |
-0.380 |
-0.178 |
| 334 |
265771_at |
AT2G48030
|
[DNAse I-like superfamily protein] |
-0.379 |
-0.067 |
| 335 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
-0.378 |
-0.055 |
| 336 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
-0.377 |
0.248 |
| 337 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.376 |
-0.561 |
| 338 |
253478_at |
AT4G32350
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.375 |
0.089 |
| 339 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
-0.374 |
-0.048 |
| 340 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.371 |
-0.234 |
| 341 |
267166_at |
AT2G37720
|
TBL15, TRICHOME BIREFRINGENCE-LIKE 15 |
-0.370 |
-0.024 |
| 342 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.368 |
-0.061 |
| 343 |
261851_at |
AT1G50460
|
ATHKL1, HKL1, hexokinase-like 1 |
-0.368 |
-0.097 |
| 344 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
-0.366 |
0.025 |
| 345 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
-0.364 |
-0.001 |
| 346 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
-0.364 |
0.118 |
| 347 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
-0.364 |
-0.060 |
| 348 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
-0.363 |
0.233 |
| 349 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.361 |
-0.238 |
| 350 |
256050_at |
AT1G07000
|
ATEXO70B2, exocyst subunit exo70 family protein B2, EXO70B2, exocyst subunit exo70 family protein B2 |
-0.361 |
-0.003 |
| 351 |
261327_at |
AT1G44830
|
[Integrase-type DNA-binding superfamily protein] |
-0.360 |
0.116 |
| 352 |
249885_at |
AT5G22940
|
F8H, FRA8 homolog |
-0.360 |
-0.084 |
| 353 |
258528_at |
AT3G06770
|
[Pectin lyase-like superfamily protein] |
-0.358 |
-0.095 |
| 354 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
-0.358 |
0.221 |
| 355 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.354 |
-0.070 |
| 356 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
-0.354 |
0.131 |
| 357 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
-0.354 |
-0.150 |
| 358 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.354 |
0.011 |
| 359 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
-0.353 |
-0.105 |
| 360 |
259987_at |
AT1G75030
|
ATLP-3, thaumatin-like protein 3, TLP-3, thaumatin-like protein 3 |
-0.352 |
0.346 |
| 361 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.348 |
0.007 |
| 362 |
261822_at |
AT1G11380
|
[PLAC8 family protein] |
-0.347 |
-0.032 |
| 363 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
-0.347 |
0.057 |
| 364 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
-0.344 |
-0.188 |
| 365 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.342 |
-0.282 |
| 366 |
253997_at |
AT4G26090
|
RPS2, RESISTANT TO P. SYRINGAE 2 |
-0.341 |
0.181 |
| 367 |
261308_at |
AT1G48480
|
RKL1, receptor-like kinase 1 |
-0.341 |
-0.032 |
| 368 |
259850_at |
AT1G72240
|
unknown |
-0.341 |
0.066 |
| 369 |
247246_at |
AT5G64620
|
ATC/VIF2, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 2, C/VIF2, cell wall / vacuolar inhibitor of fructosidase 2 |
-0.340 |
-0.032 |
| 370 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
-0.340 |
0.004 |
| 371 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
-0.340 |
-0.248 |
| 372 |
249763_at |
AT5G24010
|
[Protein kinase superfamily protein] |
-0.337 |
0.070 |
| 373 |
264361_at |
AT1G03300
|
unknown |
-0.336 |
-0.083 |
| 374 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.335 |
-0.132 |
| 375 |
253643_at |
AT4G29780
|
unknown |
-0.334 |
0.343 |
| 376 |
254574_at |
AT4G19430
|
unknown |
-0.334 |
-0.073 |
| 377 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.333 |
-0.060 |
| 378 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
-0.333 |
-0.000 |
| 379 |
250110_at |
AT5G15350
|
AtENODL17, ENODL17, early nodulin-like protein 17 |
-0.333 |
-0.032 |
| 380 |
250419_at |
AT5G11250
|
[Disease resistance protein (TIR-NBS-LRR class)] |
-0.333 |
-0.104 |
| 381 |
251861_at |
AT3G54810
|
BME3, BLUE MICROPYLAR END 3, BME3-ZF, BLUE MICROPYLAR END 3-ZINC FINGER, GATA8, GATA TRANSCRIPTION FACTOR 8 |
-0.333 |
-0.019 |
| 382 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
-0.332 |
0.006 |
| 383 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
-0.331 |
0.093 |
| 384 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
-0.330 |
-0.002 |
| 385 |
267623_at |
AT2G39650
|
unknown |
-0.329 |
0.152 |
| 386 |
251192_at |
AT3G62720
|
ATXT1, XT1, xylosyltransferase 1, XXT1, XYG XYLOSYLTRANSFERASE 1 |
-0.329 |
0.142 |
| 387 |
253925_at |
AT4G26690
|
GDPDL3, Glycerophosphodiester phosphodiesterase (GDPD) like 3, GPDL2, GLYCEROPHOSPHODIESTERASE-LIKE 2, MRH5, MUTANT ROOT HAIR 5, SHV3, SHAVEN 3 |
-0.328 |
-0.041 |
| 388 |
249918_at |
AT5G19240
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.328 |
-0.068 |
| 389 |
257636_at |
AT3G26200
|
CYP71B22, cytochrome P450, family 71, subfamily B, polypeptide 22 |
-0.326 |
0.054 |
| 390 |
260109_at |
AT1G63260
|
TET10, tetraspanin10 |
-0.324 |
-0.053 |
| 391 |
252483_at |
AT3G46600
|
[GRAS family transcription factor] |
-0.324 |
-0.128 |
| 392 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
-0.323 |
-0.052 |
| 393 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
-0.322 |
-0.048 |
| 394 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.321 |
-0.043 |
| 395 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
-0.321 |
-0.154 |
| 396 |
267381_at |
AT2G26190
|
[calmodulin-binding family protein] |
-0.320 |
-0.120 |
| 397 |
247617_at |
AT5G60270
|
LecRK-I.7, L-type lectin receptor kinase I.7 |
-0.320 |
0.009 |
| 398 |
247848_at |
AT5G58120
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.319 |
0.066 |
| 399 |
256597_at |
AT3G28500
|
[60S acidic ribosomal protein family] |
-0.319 |
0.008 |
| 400 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
-0.319 |
-0.313 |
| 401 |
263499_at |
AT2G42580
|
TTL3, tetratricopetide-repeat thioredoxin-like 3, VIT, VHI-INTERACTING TPR CONTAINING PROTEIN |
-0.317 |
-0.006 |
| 402 |
259979_at |
AT1G76600
|
unknown |
-0.316 |
-0.095 |
| 403 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
-0.315 |
-0.166 |
| 404 |
248870_at |
AT5G46710
|
[PLATZ transcription factor family protein] |
-0.315 |
0.097 |
| 405 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.314 |
0.020 |
| 406 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
-0.314 |
0.022 |
| 407 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
-0.313 |
0.024 |
| 408 |
246408_at |
AT1G57680
|
Cand1, candidate G-protein Coupled Receptor 1 |
-0.310 |
-0.049 |