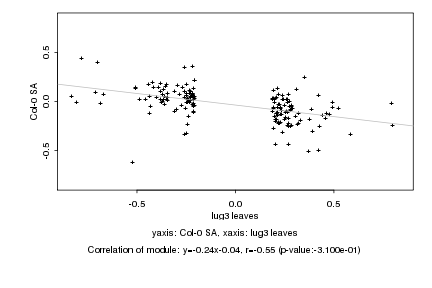
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
lug3 leaves |
Log2 signal ratio
Col-0 SA |
| 1 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.797 |
-0.237 |
| 2 |
245451_at |
AT4G16890
|
BAL, BALL, SNC1, SUPPRESSOR OF NPR1-1, CONSTITUTIVE 1 |
0.789 |
-0.012 |
| 3 |
260028_at |
AT1G29980
|
unknown |
0.584 |
-0.333 |
| 4 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.521 |
-0.070 |
| 5 |
245456_at |
AT4G16950
|
RPP5, RECOGNITION OF PERONOSPORA PARASITICA 5 |
0.492 |
-0.001 |
| 6 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.489 |
-0.057 |
| 7 |
254798_at |
AT4G13050
|
[Acyl-ACP thioesterase] |
0.473 |
-0.132 |
| 8 |
258764_at |
AT3G10720
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.461 |
-0.119 |
| 9 |
264343_at |
AT1G11850
|
unknown |
0.456 |
-0.166 |
| 10 |
248179_at |
AT5G54380
|
THE1, THESEUS1 |
0.438 |
-0.135 |
| 11 |
265656_at |
AT2G13820
|
AtXYP2, XYP2, xylogen protein 2 |
0.424 |
-0.250 |
| 12 |
251829_at |
AT3G55020
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
0.419 |
0.070 |
| 13 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
0.418 |
-0.492 |
| 14 |
260982_at |
AT1G53520
|
AtFAP3, FAP3, fatty-acid-binding protein 3 |
0.388 |
-0.297 |
| 15 |
253925_at |
AT4G26690
|
GDPDL3, Glycerophosphodiester phosphodiesterase (GDPD) like 3, GPDL2, GLYCEROPHOSPHODIESTERASE-LIKE 2, MRH5, MUTANT ROOT HAIR 5, SHV3, SHAVEN 3 |
0.386 |
-0.074 |
| 16 |
258897_at |
AT3G05730
|
[LOCATED IN: endomembrane system] |
0.372 |
-0.174 |
| 17 |
255937_at |
AT1G12610
|
DDF1, DWARF AND DELAYED FLOWERING 1 |
0.370 |
-0.500 |
| 18 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
0.347 |
0.249 |
| 19 |
265377_at |
AT2G05790
|
[O-Glycosyl hydrolases family 17 protein] |
0.330 |
-0.191 |
| 20 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
0.319 |
-0.215 |
| 21 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
0.318 |
-0.118 |
| 22 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
0.310 |
-0.232 |
| 23 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.308 |
0.130 |
| 24 |
258641_at |
AT3G08030
|
unknown |
0.304 |
-0.143 |
| 25 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
0.286 |
-0.070 |
| 26 |
257999_at |
AT3G27540
|
[beta-1,4-N-acetylglucosaminyltransferase family protein] |
0.282 |
-0.050 |
| 27 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
0.281 |
-0.031 |
| 28 |
250939_at |
AT5G03040
|
iqd2, IQ-domain 2 |
0.280 |
-0.244 |
| 29 |
259680_at |
AT1G77690
|
LAX3, like AUX1 3 |
0.279 |
-0.254 |
| 30 |
257219_at |
AT3G14930
|
HEME1 |
0.277 |
-0.076 |
| 31 |
246505_at |
AT5G16250
|
unknown |
0.275 |
-0.084 |
| 32 |
250366_at |
AT5G11420
|
unknown |
0.271 |
0.005 |
| 33 |
260944_at |
AT1G45130
|
AtBGAL5, BGAL5, beta-galactosidase 5 |
0.271 |
-0.042 |
| 34 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.269 |
0.074 |
| 35 |
264315_at |
AT1G70370
|
PG2, polygalacturonase 2 |
0.269 |
-0.169 |
| 36 |
253891_at |
AT4G27720
|
[Major facilitator superfamily protein] |
0.266 |
-0.253 |
| 37 |
262086_at |
AT1G56050
|
[GTP-binding protein-related] |
0.265 |
-0.431 |
| 38 |
250470_at |
AT5G10160
|
[Thioesterase superfamily protein] |
0.265 |
-0.222 |
| 39 |
267220_at |
AT2G02500
|
ATMEPCT, ISPD, MCT, 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLTRANSFERASE |
0.264 |
-0.109 |
| 40 |
247037_at |
AT5G67070
|
RALFL34, ralf-like 34 |
0.263 |
-0.239 |
| 41 |
266334_at |
AT2G32380
|
[Transmembrane protein 97, predicted] |
0.263 |
0.024 |
| 42 |
254737_at |
AT4G13840
|
CER26, ECERIFERUM 26 |
0.262 |
-0.001 |
| 43 |
249129_at |
AT5G43080
|
CYCA3;1, Cyclin A3;1 |
0.259 |
0.021 |
| 44 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
0.259 |
-0.096 |
| 45 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
0.254 |
-0.110 |
| 46 |
253405_at |
AT4G32800
|
[Integrase-type DNA-binding superfamily protein] |
0.251 |
-0.161 |
| 47 |
254815_at |
AT4G12420
|
SKU5 |
0.247 |
-0.183 |
| 48 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
0.244 |
-0.039 |
| 49 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.239 |
0.021 |
| 50 |
264774_at |
AT1G22890
|
unknown |
0.237 |
-0.315 |
| 51 |
254432_at |
AT4G20830
|
[FAD-binding Berberine family protein] |
0.237 |
0.028 |
| 52 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
0.234 |
0.070 |
| 53 |
259072_at |
AT3G11700
|
FLA18, FASCICLIN-like arabinogalactan protein 18 precursor |
0.231 |
-0.086 |
| 54 |
258155_at |
AT3G18130
|
RACK1C, receptor for activated C kinase 1C, RACK1C_AT, receptor for activated C kinase 1C |
0.230 |
-0.132 |
| 55 |
255714_at |
AT4G00300
|
[fringe-related protein] |
0.228 |
-0.052 |
| 56 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.228 |
-0.123 |
| 57 |
266921_at |
AT2G45970
|
CYP86A8, cytochrome P450, family 86, subfamily A, polypeptide 8, LCR, LACERATA |
0.227 |
-0.067 |
| 58 |
259592_at |
AT1G27950
|
LTPG1, glycosylphosphatidylinositol-anchored lipid protein transfer 1 |
0.225 |
-0.210 |
| 59 |
258222_at |
AT3G15680
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
0.222 |
-0.220 |
| 60 |
267355_at |
AT2G39900
|
WLIM2a, WLIM2a |
0.220 |
-0.025 |
| 61 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
0.218 |
0.074 |
| 62 |
265103_at |
AT1G31070
|
GlcNAc1pUT1, N-acetylglucosamine-1-phosphate uridylyltransferase 1 |
0.217 |
-0.029 |
| 63 |
252866_at |
AT4G39840
|
unknown |
0.215 |
-0.216 |
| 64 |
256469_at |
AT1G32540
|
LOL1, lsd one like 1 |
0.214 |
-0.213 |
| 65 |
245828_at |
AT1G57820
|
ORTH2, ORTHRUS 2, VIM1, VARIANT IN METHYLATION 1 |
0.212 |
0.135 |
| 66 |
255411_at |
AT4G03110
|
AtBRN1, AtRBP-DR1, RNA-binding protein-defense related 1, BRN1, Bruno-like 1, RBP-DR1, RNA-binding protein-defense related 1 |
0.211 |
-0.060 |
| 67 |
256983_at |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
0.210 |
-0.104 |
| 68 |
263432_at |
AT2G22230
|
[Thioesterase superfamily protein] |
0.210 |
-0.133 |
| 69 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
0.208 |
0.043 |
| 70 |
245479_at |
AT4G16140
|
[proline-rich family protein] |
0.208 |
-0.181 |
| 71 |
245002_at |
ATCG00270
|
PSBD, photosystem II reaction center protein D |
0.207 |
-0.113 |
| 72 |
249466_at |
AT5G39740
|
OLI7, OLIGOCELLULA 7, RPL5B, ribosomal protein L5 B |
0.204 |
-0.115 |
| 73 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
0.201 |
0.041 |
| 74 |
245318_at |
AT4G16980
|
[arabinogalactan-protein family] |
0.201 |
-0.174 |
| 75 |
245368_at |
AT4G15510
|
PPD1, PsbP-Domain Protein1 |
0.200 |
-0.194 |
| 76 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
0.199 |
-0.436 |
| 77 |
248074_at |
AT5G55730
|
FLA1, FASCICLIN-like arabinogalactan 1 |
0.198 |
-0.145 |
| 78 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
0.198 |
0.023 |
| 79 |
264262_at |
AT1G09200
|
H3.1, histone 3.1 |
0.198 |
-0.007 |
| 80 |
259358_at |
AT1G13250
|
GATL3, galacturonosyltransferase-like 3 |
0.193 |
0.117 |
| 81 |
261570_at |
AT1G01120
|
KCS1, 3-ketoacyl-CoA synthase 1 |
0.192 |
-0.071 |
| 82 |
247470_at |
AT5G62220
|
ATGT18, glycosyltransferase 18, GT18, glycosyltransferase 18, XLT2, Xyloglucan L-side chain galactosylTransferase position 2 |
0.191 |
0.033 |
| 83 |
258658_at |
AT3G09820
|
ADK1, adenosine kinase 1, ATADK1 |
0.191 |
-0.057 |
| 84 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
0.190 |
-0.271 |
| 85 |
249209_at |
AT5G42620
|
[metalloendopeptidases] |
0.188 |
0.039 |
| 86 |
264518_at |
AT1G09980
|
[Putative serine esterase family protein] |
0.188 |
-0.096 |
| 87 |
262708_at |
AT1G16190
|
RAD23A, RADIATION SENSITIVE23A |
0.187 |
0.027 |
| 88 |
245454_at |
AT4G16920
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.836 |
0.053 |
| 89 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.811 |
-0.008 |
| 90 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
-0.786 |
0.444 |
| 91 |
259329_at |
AT3G16360
|
AHP4, HPT phosphotransmitter 4 |
-0.712 |
0.092 |
| 92 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
-0.705 |
0.403 |
| 93 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.687 |
-0.020 |
| 94 |
254566_at |
AT4G19240
|
unknown |
-0.670 |
0.079 |
| 95 |
256789_at |
AT3G13672
|
SINA2 |
-0.527 |
-0.615 |
| 96 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.512 |
0.133 |
| 97 |
254482_at |
AT4G20370
|
TSF, TWIN SISTER OF FT |
-0.510 |
0.145 |
| 98 |
253443_at |
AT4G32551
|
LUG, LEUNIG, RON2, ROTUNDA 2 |
-0.490 |
0.025 |
| 99 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
-0.458 |
0.025 |
| 100 |
267036_at |
AT2G38465
|
unknown |
-0.444 |
0.175 |
| 101 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
-0.440 |
-0.117 |
| 102 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.439 |
0.059 |
| 103 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
-0.432 |
-0.045 |
| 104 |
254415_at |
AT4G21326
|
ATSBT3.12, subtilase 3.12, SBT3.12, subtilase 3.12 |
-0.422 |
0.203 |
| 105 |
262615_at |
AT1G13950
|
ATELF5A-1, EUKARYOTIC ELONGATION FACTOR 5A-1, EIF-5A, EIF5A, EUKARYOTIC ELONGATION FACTOR 5A, ELF5A-1, eukaryotic elongation factor 5A-1 |
-0.418 |
0.143 |
| 106 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
-0.404 |
0.049 |
| 107 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
-0.395 |
0.147 |
| 108 |
254596_at |
AT4G18975
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.381 |
0.026 |
| 109 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
-0.381 |
0.108 |
| 110 |
245302_at |
AT4G17695
|
KAN3, KANADI 3 |
-0.381 |
0.185 |
| 111 |
245310_at |
AT4G13950
|
RALFL31, ralf-like 31 |
-0.379 |
-0.003 |
| 112 |
245280_at |
AT4G16845
|
VRN2, REDUCED VERNALIZATION RESPONSE 2 |
-0.375 |
0.081 |
| 113 |
254306_at |
AT4G22330
|
ATCES1 |
-0.375 |
0.004 |
| 114 |
261222_at |
AT1G20120
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.367 |
0.027 |
| 115 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-0.366 |
0.123 |
| 116 |
250485_at |
AT5G09990
|
PROPEP5, elicitor peptide 5 precursor |
-0.363 |
0.061 |
| 117 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.363 |
-0.024 |
| 118 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
-0.357 |
0.163 |
| 119 |
247212_at |
AT5G65040
|
unknown |
-0.353 |
0.175 |
| 120 |
266480_at |
AT2G31130
|
unknown |
-0.351 |
0.030 |
| 121 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.350 |
0.014 |
| 122 |
246968_at |
AT5G24870
|
[RING/U-box superfamily protein] |
-0.350 |
0.049 |
| 123 |
260933_at |
AT1G02470
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.345 |
0.091 |
| 124 |
266355_at |
AT2G01400
|
unknown |
-0.314 |
-0.092 |
| 125 |
264522_at |
AT1G10050
|
[glycosyl hydrolase family 10 protein / carbohydrate-binding domain-containing protein] |
-0.312 |
0.110 |
| 126 |
251012_at |
AT5G02580
|
unknown |
-0.300 |
-0.074 |
| 127 |
251191_at |
AT3G62590
|
[alpha/beta-Hydrolases superfamily protein] |
-0.298 |
0.165 |
| 128 |
248994_at |
AT5G45250
|
RPS4, RESISTANT TO P. SYRINGAE 4 |
-0.287 |
0.078 |
| 129 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
-0.277 |
-0.035 |
| 130 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.271 |
0.143 |
| 131 |
261272_at |
AT1G26665
|
[Mediator complex, subunit Med10] |
-0.263 |
0.044 |
| 132 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
-0.262 |
0.351 |
| 133 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.262 |
-0.331 |
| 134 |
262918_at |
AT1G65000
|
unknown |
-0.259 |
0.108 |
| 135 |
255487_at |
AT4G02610
|
[Aldolase-type TIM barrel family protein] |
-0.257 |
-0.068 |
| 136 |
253640_at |
AT4G30630
|
unknown |
-0.253 |
0.179 |
| 137 |
247063_at |
AT5G66820
|
unknown |
-0.252 |
-0.006 |
| 138 |
245331_at |
AT4G14410
|
bHLH104, basic Helix-Loop-Helix 104 |
-0.252 |
0.062 |
| 139 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.251 |
-0.319 |
| 140 |
266839_at |
AT2G25930
|
ELF3, EARLY FLOWERING 3, PYK20 |
-0.249 |
0.086 |
| 141 |
252888_at |
AT4G39210
|
APL3 |
-0.248 |
0.031 |
| 142 |
254965_at |
AT4G11090
|
TBL23, TRICHOME BIREFRINGENCE-LIKE 23 |
-0.246 |
-0.008 |
| 143 |
262905_at |
AT1G59730
|
ATH7, thioredoxin H-type 7, TH7, thioredoxin H-type 7 |
-0.246 |
-0.231 |
| 144 |
258756_at |
AT3G11960
|
[Cleavage and polyadenylation specificity factor (CPSF) A subunit protein] |
-0.244 |
0.030 |
| 145 |
251929_at |
AT3G53920
|
SIG3, SIGMA FACTOR 3, SIGC, RNApolymerase sigma-subunit C |
-0.241 |
-0.148 |
| 146 |
254350_at |
AT4G22280
|
[F-box/RNI-like superfamily protein] |
-0.241 |
0.003 |
| 147 |
264091_at |
AT1G79110
|
BRG2, BOI-related gene 2 |
-0.238 |
0.117 |
| 148 |
247514_at |
AT5G61640
|
ATMSRA1, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE A1, PMSR1, peptidemethionine sulfoxide reductase 1 |
-0.238 |
-0.004 |
| 149 |
251644_at |
AT3G57540
|
[Remorin family protein] |
-0.234 |
0.085 |
| 150 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
-0.233 |
0.014 |
| 151 |
258312_at |
AT3G16170
|
AAE13, acyl activating enzyme 13 |
-0.231 |
0.049 |
| 152 |
257369_at |
AT2G35550
|
ATBPC7, BASIC PENTACYSTEINE 7, BBR, BPC7, basic pentacysteine 7 |
-0.229 |
0.107 |
| 153 |
251393_at |
AT3G60640
|
ATG8G, AUTOPHAGY 8G |
-0.229 |
0.060 |
| 154 |
246909_at |
AT5G25770
|
[alpha/beta-Hydrolases superfamily protein] |
-0.223 |
0.090 |
| 155 |
256220_at |
AT1G56230
|
unknown |
-0.223 |
0.071 |
| 156 |
254253_at |
AT4G23320
|
CRK24, cysteine-rich RLK (RECEPTOR-like protein kinase) 24 |
-0.223 |
0.061 |
| 157 |
257580_at |
AT3G06210
|
[ARM repeat superfamily protein] |
-0.222 |
0.091 |
| 158 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
-0.221 |
-0.030 |
| 159 |
250597_at |
AT5G07680
|
ANAC079, Arabidopsis NAC domain containing protein 79, ANAC080, NAC domain containing protein 80, ATNAC4, NAC080, NAC domain containing protein 80, NAC4 |
-0.219 |
0.366 |
| 160 |
253236_at |
AT4G34370
|
ARI1, ARIADNE 1, ATARI1, ARABIDOPSIS ARIADNE 1 |
-0.219 |
0.076 |
| 161 |
246262_at |
AT1G31790
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.218 |
-0.046 |
| 162 |
256061_at |
AT1G07040
|
unknown |
-0.216 |
0.022 |
| 163 |
262888_at |
AT1G14790
|
AtRDR1, ATRDRP1, RDR1, RNA-dependent RNA polymerase 1 |
-0.216 |
0.139 |
| 164 |
250487_at |
AT5G09690
|
ATMGT7, ARABIDOPSIS THALIANA MAGNESIUM TRANSPORTER 7, MGT7, magnesium transporter 7, MRS2-7 |
-0.216 |
-0.034 |
| 165 |
262640_at |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.216 |
-0.014 |
| 166 |
251175_at |
AT3G63250
|
ATHMT-2, HOMOCYSTEINE METHYLTRANSFERASE-2, HMT-2, HOMOCYSTEINE METHYLTRANSFERASE-2, HMT2, homocysteine methyltransferase 2 |
-0.215 |
-0.110 |
| 167 |
257923_at |
AT3G23160
|
unknown |
-0.214 |
-0.094 |
| 168 |
261145_at |
AT1G19730
|
ATH4, thioredoxin H-type 4, ATTRX4 |
-0.214 |
0.045 |
| 169 |
247835_at |
AT5G57910
|
unknown |
-0.212 |
0.052 |
| 170 |
245390_at |
AT4G17650
|
[Polyketide cyclase / dehydrase and lipid transport protein] |
-0.211 |
0.037 |
| 171 |
264405_at |
AT1G10210
|
ATMPK1, mitogen-activated protein kinase 1, MPK1, mitogen-activated protein kinase 1 |
-0.210 |
0.222 |
| 172 |
261845_at |
AT1G15960
|
ATNRAMP6, NRAMP6, NRAMP metal ion transporter 6 |
-0.210 |
-0.040 |
| 173 |
250363_at |
AT5G11390
|
WIT1, WPP domain-interacting protein 1 |
-0.210 |
0.221 |