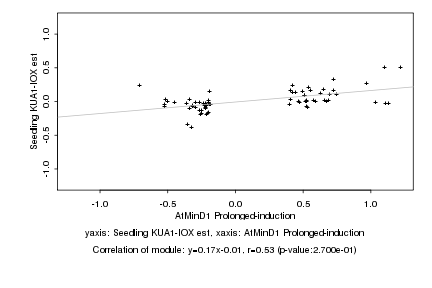
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
AtMinD1 Prolonged-induction |
Log2 signal ratio
Seedling KUA1-IOX est |
| 1 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
1.216 |
0.515 |
| 2 |
246380_at |
AT1G57750
|
CYP96A15, cytochrome P450, family 96, subfamily A, polypeptide 15, MAH1, MID-CHAIN ALKANE HYDROXYLASE 1 |
1.125 |
-0.016 |
| 3 |
249939_at |
AT5G22430
|
[Pollen Ole e 1 allergen and extensin family protein] |
1.103 |
-0.018 |
| 4 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
1.099 |
0.507 |
| 5 |
250610_at |
AT5G07550
|
ATGRP19, GRP19, glycine-rich protein 19 |
1.028 |
-0.009 |
| 6 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.964 |
0.279 |
| 7 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.746 |
0.108 |
| 8 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.722 |
0.176 |
| 9 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.719 |
0.330 |
| 10 |
253012_at |
AT4G37900
|
unknown |
0.694 |
0.116 |
| 11 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.682 |
0.026 |
| 12 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
0.667 |
0.008 |
| 13 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.651 |
0.029 |
| 14 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.650 |
0.186 |
| 15 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.622 |
0.119 |
| 16 |
265441_at |
AT2G20870
|
[cell wall protein precursor, putative] |
0.588 |
0.002 |
| 17 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.570 |
0.025 |
| 18 |
258203_at |
AT3G13950
|
unknown |
0.552 |
0.172 |
| 19 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.537 |
0.215 |
| 20 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.532 |
-0.077 |
| 21 |
262697_at |
AT1G75940
|
ATA27, BGLU20, BETA GLUCOSIDASE 20 |
0.521 |
0.010 |
| 22 |
248496_at |
AT5G50790
|
AtSWEET10, SWEET10 |
0.518 |
-0.070 |
| 23 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.518 |
0.019 |
| 24 |
258962_at |
AT3G10570
|
CYP77A6, cytochrome P450, family 77, subfamily A, polypeptide 6 |
0.513 |
0.003 |
| 25 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.509 |
0.100 |
| 26 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
0.507 |
0.101 |
| 27 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.488 |
0.153 |
| 28 |
264774_at |
AT1G22890
|
unknown |
0.469 |
-0.014 |
| 29 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.460 |
0.010 |
| 30 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.443 |
0.136 |
| 31 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.421 |
0.243 |
| 32 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.420 |
0.147 |
| 33 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
0.404 |
0.171 |
| 34 |
259340_at |
AT3G03870
|
unknown |
0.404 |
0.041 |
| 35 |
261772_at |
AT1G76240
|
unknown |
0.396 |
-0.036 |
| 36 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
-0.712 |
0.245 |
| 37 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.527 |
-0.071 |
| 38 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
-0.524 |
-0.031 |
| 39 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.519 |
0.032 |
| 40 |
249302_at |
AT5G41450
|
[RING/U-box superfamily protein] |
-0.505 |
0.009 |
| 41 |
250142_at |
AT5G14650
|
[Pectin lyase-like superfamily protein] |
-0.457 |
-0.008 |
| 42 |
264640_at |
AT1G65680
|
ATEXPB2, expansin B2, ATHEXP BETA 1.4, EXPB2, expansin B2 |
-0.366 |
-0.023 |
| 43 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-0.357 |
-0.337 |
| 44 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.341 |
0.032 |
| 45 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
-0.340 |
-0.098 |
| 46 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.328 |
-0.376 |
| 47 |
255549_at |
AT4G01950
|
ATGPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3, GPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3 |
-0.324 |
-0.067 |
| 48 |
266941_at |
AT2G18980
|
[Peroxidase superfamily protein] |
-0.323 |
-0.055 |
| 49 |
265029_at |
AT1G24625
|
ZFP7, zinc finger protein 7 |
-0.300 |
-0.005 |
| 50 |
248380_at |
AT5G51820
|
ATPGMP, ARABIDOPSIS THALIANA PHOSPHOGLUCOMUTASE, PGM, phosphoglucomutase, PGM1, STF1, STARCH-FREE 1 |
-0.297 |
-0.076 |
| 51 |
266961_at |
AT2G07720
|
[gypsy-like retrotransposon family (Athila), has a 1.1e-59 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
-0.268 |
-0.005 |
| 52 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
-0.268 |
-0.133 |
| 53 |
245966_at |
AT5G19790
|
RAP2.11, related to AP2 11 |
-0.265 |
-0.183 |
| 54 |
245555_at |
AT4G15390
|
[HXXXD-type acyl-transferase family protein] |
-0.258 |
-0.163 |
| 55 |
257697_at |
AT3G12700
|
NANA, NANA |
-0.253 |
-0.123 |
| 56 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
-0.243 |
-0.020 |
| 57 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.232 |
-0.053 |
| 58 |
257533_at |
AT3G10840
|
[alpha/beta-Hydrolases superfamily protein] |
-0.228 |
-0.089 |
| 59 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.227 |
-0.081 |
| 60 |
255126_at |
AT4G08270
|
unknown |
-0.224 |
-0.015 |
| 61 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.216 |
-0.055 |
| 62 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-0.214 |
-0.192 |
| 63 |
248920_at |
AT5G45930
|
CHL I2, CHLI-2, CHLI2, magnesium chelatase i2 |
-0.210 |
-0.171 |
| 64 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.202 |
-0.158 |
| 65 |
258032_at |
AT3G21170
|
[F-box and associated interaction domains-containing protein] |
-0.199 |
-0.011 |
| 66 |
257135_at |
AT3G12900
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.199 |
0.029 |
| 67 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
-0.198 |
0.162 |
| 68 |
260030_at |
AT1G68880
|
AtbZIP, basic leucine-zipper 8, bZIP, basic leucine-zipper 8 |
-0.196 |
-0.043 |
| 69 |
256890_at |
AT3G23830
|
AtGRP4, GR-RBP4, GRP4, glycine-rich RNA-binding protein 4 |
-0.191 |
-0.037 |