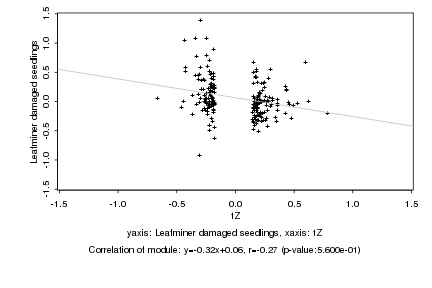
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
tZ |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
0.779 |
-0.188 |
| 2 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
0.622 |
0.003 |
| 3 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
0.589 |
0.668 |
| 4 |
250223_at |
AT5G14070
|
ROXY2 |
0.522 |
-0.033 |
| 5 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
0.493 |
-0.062 |
| 6 |
266321_at |
AT2G46660
|
CYP78A6, cytochrome P450, family 78, subfamily A, polypeptide 6, EOD3, enhancer of da1-1 |
0.477 |
-0.287 |
| 7 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
0.460 |
-0.049 |
| 8 |
252070_at |
AT3G51680
|
AtSDR2, SDR2, short-chain dehydrogenase/reductase 2 |
0.451 |
-0.008 |
| 9 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
0.433 |
0.221 |
| 10 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
0.429 |
0.198 |
| 11 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.426 |
0.259 |
| 12 |
250413_at |
AT5G11160
|
APT5, adenine phosphoribosyltransferase 5 |
0.418 |
-0.189 |
| 13 |
247956_at |
AT5G56970
|
ATCKX3, CKX3, cytokinin oxidase 3 |
0.357 |
0.045 |
| 14 |
252501_at |
AT3G46880
|
unknown |
0.357 |
-0.024 |
| 15 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
0.356 |
-0.052 |
| 16 |
257288_at |
AT3G29670
|
PMAT2, phenolic glucoside malonyltransferase 2 |
0.354 |
-0.138 |
| 17 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
0.344 |
-0.339 |
| 18 |
246395_at |
AT1G58170
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.338 |
-0.262 |
| 19 |
247991_at |
AT5G56320
|
ATEXP14, ATEXPA14, expansin A14, ATHEXP ALPHA 1.5, EXP14, EXPANSIN 14, EXPA14, expansin A14 |
0.314 |
0.029 |
| 20 |
261369_at |
AT1G53060
|
[Legume lectin family protein] |
0.308 |
0.053 |
| 21 |
262704_at |
AT1G16530
|
ASL9, ASYMMETRIC LEAVES 2-like 9, LBD3, LOB DOMAIN-CONTAINING PROTEIN 3 |
0.300 |
-0.047 |
| 22 |
267058_at |
AT2G32510
|
MAPKKK17, mitogen-activated protein kinase kinase kinase 17 |
0.297 |
0.552 |
| 23 |
264790_at |
AT2G17820
|
AHK1, ATHK1, histidine kinase 1, HK1, histidine kinase 1 |
0.296 |
-0.079 |
| 24 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
0.286 |
0.076 |
| 25 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.281 |
0.402 |
| 26 |
250908_at |
AT5G03680
|
PTL, PETAL LOSS |
0.280 |
0.012 |
| 27 |
252536_at |
AT3G45700
|
[Major facilitator superfamily protein] |
0.273 |
0.021 |
| 28 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
0.273 |
-0.414 |
| 29 |
266439_s_at |
AT2G43200
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.269 |
-0.141 |
| 30 |
257353_at |
AT2G23050
|
MEL4, MAB4/ENP/NPY1-LIKE 4, NPY4, NAKED PINS IN YUC MUTANTS 4 |
0.266 |
0.010 |
| 31 |
259346_at |
AT3G03910
|
glutamate dehydrogenase 3 |
0.263 |
-0.287 |
| 32 |
247483_at |
AT5G62420
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.261 |
-0.005 |
| 33 |
253100_at |
AT4G37400
|
CYP81F3, cytochrome P450, family 81, subfamily F, polypeptide 3 |
0.257 |
-0.053 |
| 34 |
265967_at |
AT2G37450
|
UMAMIT13, Usually multiple acids move in and out Transporters 13 |
0.250 |
-0.315 |
| 35 |
260484_at |
AT1G68360
|
[C2H2 and C2HC zinc fingers superfamily protein] |
0.250 |
0.086 |
| 36 |
261934_at |
AT1G22400
|
ATUGT85A1, ARABIDOPSIS THALIANA UDP-GLUCOSYL TRANSFERASE 85A1, UGT85A1 |
0.244 |
0.249 |
| 37 |
267327_at |
AT2G19410
|
[U-box domain-containing protein kinase family protein] |
0.244 |
0.023 |
| 38 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
0.244 |
0.332 |
| 39 |
252626_at |
AT3G44940
|
unknown |
0.244 |
-0.172 |
| 40 |
254632_at |
AT4G18630
|
unknown |
0.235 |
0.323 |
| 41 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
0.232 |
-0.321 |
| 42 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
0.230 |
-0.204 |
| 43 |
263382_at |
AT2G40230
|
[HXXXD-type acyl-transferase family protein] |
0.229 |
0.196 |
| 44 |
263932_at |
AT2G35990
|
LOG2, LONELY GUY 2 |
0.228 |
0.009 |
| 45 |
251062_at |
AT5G01840
|
ATOFP1, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 1, OFP1, ovate family protein 1 |
0.222 |
0.324 |
| 46 |
253478_at |
AT4G32350
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.222 |
-0.270 |
| 47 |
248643_at |
AT5G49130
|
[MATE efflux family protein] |
0.220 |
-0.003 |
| 48 |
259042_at |
AT3G03450
|
RGL2, RGA-like 2 |
0.218 |
-0.341 |
| 49 |
264501_at |
AT1G09390
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.213 |
-0.173 |
| 50 |
247005_at |
AT5G67520
|
adenosine-5'-phosphosulfate (APS) kinase 4 |
0.209 |
0.149 |
| 51 |
247399_at |
AT5G62960
|
unknown |
0.207 |
0.024 |
| 52 |
251374_at |
AT3G60390
|
HAT3, homeobox-leucine zipper protein 3 |
0.207 |
-0.256 |
| 53 |
260539_at |
AT2G43480
|
[Peroxidase superfamily protein] |
0.202 |
0.043 |
| 54 |
247467_at |
AT5G62130
|
[Per1-like family protein] |
0.202 |
-0.205 |
| 55 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.201 |
0.162 |
| 56 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.199 |
0.150 |
| 57 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
0.199 |
-0.215 |
| 58 |
256615_at |
AT3G22250
|
[UDP-Glycosyltransferase superfamily protein] |
0.198 |
-0.018 |
| 59 |
256508_at |
AT1G75140
|
unknown |
0.193 |
0.117 |
| 60 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
0.192 |
-0.318 |
| 61 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
0.189 |
-0.224 |
| 62 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
0.188 |
-0.500 |
| 63 |
253851_at |
AT4G28110
|
AtMYB41, myb domain protein 41, MYB41, myb domain protein 41 |
0.188 |
0.091 |
| 64 |
250205_at |
AT5G14020
|
[Endosomal targeting BRO1-like domain-containing protein] |
0.187 |
0.044 |
| 65 |
266008_at |
AT2G37390
|
NAKR2, SODIUM POTASSIUM ROOT DEFECTIVE 2 |
0.186 |
-0.105 |
| 66 |
266770_at |
AT2G03090
|
ATEXP15, ATEXPA15, expansin A15, ATHEXP ALPHA 1.3, EXP15, EXPANSIN 15, EXPA15, expansin A15 |
0.185 |
-0.090 |
| 67 |
250396_at |
AT5G10970
|
[C2H2 and C2HC zinc fingers superfamily protein] |
0.185 |
-0.316 |
| 68 |
260528_at |
AT2G47260
|
ATWRKY23, WRKY DNA-BINDING PROTEIN 23, WRKY23, WRKY DNA-binding protein 23 |
0.184 |
0.082 |
| 69 |
249484_at |
AT5G38970
|
ATBR6OX, BR6OX, BRASSINOSTEROID-6-OXIDASE, BR6OX1, brassinosteroid-6-oxidase 1, CYP85A1 |
0.184 |
-0.104 |
| 70 |
247769_at |
AT5G58910
|
LAC16, laccase 16 |
0.183 |
0.010 |
| 71 |
253686_at |
AT4G29750
|
[CRS1 / YhbY (CRM) domain-containing protein] |
0.181 |
-0.043 |
| 72 |
245133_at |
AT2G45310
|
GAE4, UDP-D-glucuronate 4-epimerase 4 |
0.180 |
-0.185 |
| 73 |
266175_at |
AT2G02450
|
ANAC034, Arabidopsis NAC domain containing protein 34, ANAC035, NAC domain containing protein 35, AtLOV1, LOV1, LONG VEGETATIVE PHASE 1, NAC035, NAC domain containing protein 35 |
0.180 |
0.328 |
| 74 |
247159_at |
AT5G65800
|
ACS5, ACC synthase 5, ATACS5, CIN5, CYTOKININ-INSENSITIVE 5, ETO2, ETHYLENE OVERPRODUCER 2 |
0.180 |
-0.006 |
| 75 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
0.179 |
0.519 |
| 76 |
256650_at |
AT3G13620
|
PUT4, POLYAMINE UPTAKE TRANSPORTER 4 |
0.178 |
0.011 |
| 77 |
256713_at |
AT2G34060
|
[Peroxidase superfamily protein] |
0.177 |
-0.143 |
| 78 |
263236_at |
AT1G10470
|
ARR4, response regulator 4, ATRR1, RESPONCE REGULATOR 1, IBC7, INDUCED BY CYTOKININ 7, MEE7, maternal effect embryo arrest 7 |
0.175 |
-0.061 |
| 79 |
260053_at |
AT1G78120
|
TPR12, tetratricopeptide repeat 12 |
0.174 |
-0.050 |
| 80 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
0.173 |
-0.373 |
| 81 |
257460_at |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
0.172 |
0.418 |
| 82 |
256293_at |
AT1G69440
|
AGO7, ARGONAUTE7, ZIP, ZIPPY |
0.171 |
-0.256 |
| 83 |
248205_at |
AT5G54300
|
unknown |
0.171 |
0.559 |
| 84 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.169 |
0.433 |
| 85 |
247340_at |
AT5G63700
|
[zinc ion binding] |
0.167 |
-0.297 |
| 86 |
254453_at |
AT4G21120
|
AAT1, amino acid transporter 1, CAT1, CATIONIC AMINO ACID TRANSPORTER 1 |
0.164 |
0.419 |
| 87 |
252543_at |
AT3G45780
|
JK224, NPH1, NONPHOTOTROPIC HYPOCOTYL 1, PHOT1, phototropin 1, RPT1, ROOT PHOTOTROPISM 1 |
0.163 |
-0.098 |
| 88 |
249408_at |
AT5G40330
|
ATMYB23, MYB DOMAIN PROTEIN 23, ATMYBRTF, MYB23, myb domain protein 23 |
0.163 |
-0.044 |
| 89 |
253692_at |
AT4G29720
|
ATPAO5, polyamine oxidase 5, PAO5, polyamine oxidase 5 |
0.161 |
-0.225 |
| 90 |
261910_at |
AT1G80730
|
ATZFP1, ARABIDOPSIS THALIANA ZINC-FINGER PROTEIN 1, ZFP1, zinc-finger protein 1 |
0.161 |
-0.045 |
| 91 |
255676_at |
AT4G00490
|
BAM2, beta-amylase 2, BMY9, BETA-AMYLASE 9 |
0.160 |
-0.266 |
| 92 |
257976_at |
AT3G20840
|
PLT1, PLETHORA 1 |
0.158 |
0.015 |
| 93 |
256066_at |
AT1G06980
|
unknown |
0.157 |
-0.405 |
| 94 |
259677_at |
AT1G77740
|
PIP5K2, phosphatidylinositol-4-phosphate 5-kinase 2 |
0.157 |
-0.143 |
| 95 |
249566_at |
AT5G38450
|
CYP735A1, cytochrome P450, family 735, subfamily A, polypeptide 1 |
0.156 |
0.063 |
| 96 |
264804_at |
AT1G08590
|
[Leucine-rich receptor-like protein kinase family protein] |
0.154 |
-0.053 |
| 97 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.153 |
-0.088 |
| 98 |
249971_at |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
0.152 |
0.675 |
| 99 |
255939_at |
AT1G12730
|
[GPI transamidase subunit PIG-U] |
0.151 |
-0.088 |
| 100 |
262881_at |
AT1G64890
|
[Major facilitator superfamily protein] |
0.151 |
0.325 |
| 101 |
250613_at |
AT5G07240
|
IQD24, IQ-domain 24 |
0.149 |
-0.474 |
| 102 |
247741_at |
AT5G58960
|
GIL1, GRAVITROPIC IN THE LIGHT |
0.149 |
-0.330 |
| 103 |
250504_at |
AT5G09840
|
[Putative endonuclease or glycosyl hydrolase] |
0.149 |
-0.104 |
| 104 |
251665_at |
AT3G57040
|
ARR9, response regulator 9, ATRR4, RESPONSE REGULATOR 4 |
0.148 |
-0.348 |
| 105 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.148 |
0.509 |
| 106 |
247707_at |
AT5G59450
|
[GRAS family transcription factor] |
0.147 |
0.089 |
| 107 |
253794_at |
AT4G28720
|
YUC8, YUCCA 8 |
0.147 |
-0.034 |
| 108 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
0.144 |
-0.175 |
| 109 |
249798_at |
AT5G23730
|
EFO2, EARLY FLOWERING BY OVEREXPRESSION 2, RUP2, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 2 |
0.143 |
-0.299 |
| 110 |
249364_at |
AT5G40590
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.671 |
0.056 |
| 111 |
254037_at |
AT4G25760
|
ATGDU2, glutamine dumper 2, GDU2, glutamine dumper 2 |
-0.465 |
-0.090 |
| 112 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
-0.443 |
0.007 |
| 113 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.439 |
1.055 |
| 114 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
-0.428 |
0.598 |
| 115 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
-0.427 |
0.524 |
| 116 |
258570_at |
AT3G04530
|
ATPPCK2, PEPCK2, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 2, PPCK2, phosphoenolpyruvate carboxylase kinase 2 |
-0.372 |
-0.209 |
| 117 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.368 |
0.105 |
| 118 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
-0.345 |
1.092 |
| 119 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
-0.342 |
0.461 |
| 120 |
266974_at |
AT2G39370
|
MAKR4, MEMBRANE-ASSOCIATED KINASE REGULATOR 4 |
-0.340 |
-0.037 |
| 121 |
245076_at |
AT2G23170
|
GH3.3 |
-0.336 |
0.781 |
| 122 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
-0.320 |
0.453 |
| 123 |
247205_at |
AT5G64890
|
PROPEP2, elicitor peptide 2 precursor |
-0.316 |
0.392 |
| 124 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
-0.316 |
0.130 |
| 125 |
265856_at |
AT2G42430
|
ASL18, ASYMMETRIC LEAVES2-LIKE 18, LBD16, lateral organ boundaries-domain 16 |
-0.310 |
-0.014 |
| 126 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
-0.307 |
0.477 |
| 127 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.307 |
-0.918 |
| 128 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
-0.306 |
0.061 |
| 129 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
-0.305 |
0.595 |
| 130 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
-0.304 |
1.403 |
| 131 |
257927_at |
AT3G23240
|
ATERF1, ETHYLENE RESPONSE FACTOR 1, ERF1, ethylene response factor 1 |
-0.293 |
0.360 |
| 132 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
-0.293 |
0.212 |
| 133 |
249522_at |
AT5G38700
|
unknown |
-0.292 |
0.219 |
| 134 |
250582_at |
AT5G07580
|
[Integrase-type DNA-binding superfamily protein] |
-0.282 |
-0.145 |
| 135 |
257375_at |
AT2G38640
|
unknown |
-0.277 |
0.377 |
| 136 |
265245_at |
AT2G43060
|
AtIBH1, IBH1, ILI1 binding bHLH 1 |
-0.273 |
0.207 |
| 137 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
-0.272 |
0.001 |
| 138 |
255945_at |
AT5G28610
|
unknown |
-0.270 |
0.104 |
| 139 |
267077_at |
AT2G40970
|
MYBC1 |
-0.265 |
-0.103 |
| 140 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
-0.264 |
0.361 |
| 141 |
263330_at |
AT2G15320
|
[Leucine-rich repeat (LRR) family protein] |
-0.261 |
-0.138 |
| 142 |
251197_at |
AT3G62960
|
[Thioredoxin superfamily protein] |
-0.260 |
0.043 |
| 143 |
260144_at |
AT1G71960
|
ABCG25, ATP-binding casette G25, ATABCG25, Arabidopsis thaliana ATP-binding cassette G25 |
-0.258 |
0.053 |
| 144 |
250051_at |
AT5G17800
|
AtMYB56, myb domain protein 56, MYB56, myb domain protein 56 |
-0.257 |
-0.004 |
| 145 |
265539_at |
AT2G15830
|
unknown |
-0.253 |
0.140 |
| 146 |
260668_at |
AT1G19530
|
unknown |
-0.252 |
0.053 |
| 147 |
245906_at |
AT5G11070
|
unknown |
-0.251 |
-0.016 |
| 148 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
-0.250 |
0.793 |
| 149 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
-0.249 |
1.085 |
| 150 |
251565_at |
AT3G58190
|
ASL16, ASYMMETRIC LEAVES 2-LIKE 16, LBD29, lateral organ boundaries-domain 29 |
-0.249 |
-0.007 |
| 151 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
-0.249 |
0.144 |
| 152 |
265769_at |
AT2G48090
|
unknown |
-0.247 |
-0.025 |
| 153 |
251218_at |
AT3G62410
|
CP12, CP12 DOMAIN-CONTAINING PROTEIN 1, CP12-2, CP12 domain-containing protein 2 |
-0.244 |
-0.168 |
| 154 |
255937_at |
AT1G12610
|
DDF1, DWARF AND DELAYED FLOWERING 1 |
-0.241 |
0.601 |
| 155 |
256537_at |
AT1G33340
|
[ENTH/ANTH/VHS superfamily protein] |
-0.240 |
-0.107 |
| 156 |
259371_at |
AT1G69080
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.240 |
-0.214 |
| 157 |
256137_at |
AT1G48690
|
[Auxin-responsive GH3 family protein] |
-0.238 |
-0.056 |
| 158 |
248770_at |
AT5G47740
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.236 |
0.238 |
| 159 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
-0.234 |
0.053 |
| 160 |
258993_at |
AT3G08940
|
LHCB4.2, light harvesting complex photosystem II |
-0.234 |
-0.064 |
| 161 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
-0.229 |
0.525 |
| 162 |
245445_at |
AT4G16750
|
[Integrase-type DNA-binding superfamily protein] |
-0.229 |
0.155 |
| 163 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
-0.229 |
0.141 |
| 164 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
-0.229 |
-0.048 |
| 165 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
-0.228 |
0.709 |
| 166 |
262286_at |
AT1G68585
|
unknown |
-0.228 |
-0.401 |
| 167 |
255576_at |
AT4G01440
|
UMAMIT31, Usually multiple acids move in and out Transporters 31 |
-0.227 |
-0.088 |
| 168 |
263892_at |
AT2G36890
|
ATMYB38, MYB DOMAIN PROTEIN 38, BIT1, BLUE INSENSITIVE TRAIT 1, MYB38, MYB DOMAIN PROTEIN 38, RAX2, REGULATOR OF AXILLARY MERISTEMS 2 |
-0.225 |
0.015 |
| 169 |
250002_at |
AT5G18690
|
AGP25, arabinogalactan protein 25, ATAGP25, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEINS 25 |
-0.224 |
-0.409 |
| 170 |
255028_at |
AT4G09890
|
unknown |
-0.223 |
-0.490 |
| 171 |
249941_at |
AT5G22270
|
unknown |
-0.219 |
0.470 |
| 172 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
-0.218 |
0.303 |
| 173 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
-0.218 |
0.114 |
| 174 |
250098_at |
AT5G17350
|
unknown |
-0.217 |
0.193 |
| 175 |
256720_at |
AT2G34140
|
[Dof-type zinc finger DNA-binding family protein] |
-0.212 |
0.101 |
| 176 |
261769_at |
AT1G76100
|
PETE1, plastocyanin 1 |
-0.211 |
-0.077 |
| 177 |
263126_at |
AT1G78460
|
[SOUL heme-binding family protein] |
-0.209 |
0.324 |
| 178 |
260167_at |
AT1G71970
|
unknown |
-0.207 |
-0.278 |
| 179 |
263438_at |
AT2G28660
|
[Chloroplast-targeted copper chaperone protein] |
-0.207 |
0.067 |
| 180 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
-0.205 |
0.406 |
| 181 |
251423_at |
AT3G60550
|
CYCP3;2, cyclin p3;2 |
-0.205 |
-0.042 |
| 182 |
245885_at |
AT5G09440
|
EXL4, EXORDIUM like 4 |
-0.204 |
0.481 |
| 183 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
-0.204 |
0.254 |
| 184 |
262703_at |
AT1G16510
|
SAUR41, SMALL AUXIN UPREGULATED 41 |
-0.204 |
0.263 |
| 185 |
264130_at |
AT1G79160
|
unknown |
-0.203 |
0.068 |
| 186 |
251940_at |
AT3G53450
|
LOG4, LONELY GUY 4 |
-0.199 |
0.078 |
| 187 |
260608_at |
AT2G43870
|
[Pectin lyase-like superfamily protein] |
-0.199 |
0.003 |
| 188 |
252232_at |
AT3G49760
|
AtbZIP5, basic leucine-zipper 5, bZIP5, basic leucine-zipper 5 |
-0.198 |
0.059 |
| 189 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.198 |
-0.334 |
| 190 |
267069_at |
AT2G41010
|
ATCAMBP25, calmodulin (CAM)-binding protein of 25 kDa, CAMBP25, calmodulin (CAM)-binding protein of 25 kDa |
-0.197 |
-0.024 |
| 191 |
257690_at |
AT3G12830
|
SAUR72, SMALL AUXIN UPREGULATED 72 |
-0.196 |
0.256 |
| 192 |
249972_at |
AT5G19040
|
ATIPT5, Arabidopsis thaliana ISOPENTENYLTRANSFERASE 5, IPT5, isopentenyltransferase 5 |
-0.196 |
-0.013 |
| 193 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
-0.196 |
0.177 |
| 194 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
-0.195 |
0.489 |
| 195 |
264696_at |
AT1G70230
|
AXY4, ALTERED XYLOGLUCAN 4, TBL27, TRICHOME BIREFRINGENCE-LIKE 27 |
-0.193 |
-0.065 |
| 196 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
-0.192 |
-0.178 |
| 197 |
245731_at |
AT1G73500
|
ATMKK9, MKK9, MAP kinase kinase 9 |
-0.192 |
0.443 |
| 198 |
262170_at |
AT1G74940
|
unknown |
-0.191 |
-0.139 |
| 199 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
-0.191 |
0.283 |
| 200 |
246390_at |
AT1G77330
|
ACO5, ACC oxidase 5 |
-0.191 |
-0.124 |
| 201 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
-0.190 |
0.387 |
| 202 |
250231_at |
AT5G13910
|
LEP, LEAFY PETIOLE |
-0.190 |
-0.006 |
| 203 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
-0.190 |
0.441 |
| 204 |
258560_at |
AT3G06020
|
FAF4, FANTASTIC FOUR 4 |
-0.190 |
0.226 |
| 205 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
-0.189 |
0.902 |
| 206 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
-0.188 |
0.029 |
| 207 |
266546_at |
AT2G35270
|
AHL21, AT-hook motif nuclear localized protein 21, GIK, GIANT KILLER |
-0.188 |
-0.018 |
| 208 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.186 |
-0.620 |
| 209 |
254178_at |
AT4G23880
|
unknown |
-0.185 |
0.289 |
| 210 |
266824_at |
AT2G22800
|
HAT9 |
-0.183 |
-0.042 |
| 211 |
252173_at |
AT3G50650
|
[GRAS family transcription factor] |
-0.183 |
0.240 |
| 212 |
256097_at |
AT1G13670
|
unknown |
-0.182 |
-0.005 |
| 213 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
-0.180 |
0.225 |
| 214 |
247038_at |
AT5G67160
|
EPS1, ENHANCED PSEUDOMONAS SUSCEPTIBILTY 1 |
-0.179 |
-0.048 |
| 215 |
264697_at |
AT1G70210
|
ATCYCD1;1, CYCD1;1, CYCLIN D1;1 |
-0.179 |
-0.442 |
| 216 |
264889_at |
AT1G23050
|
hydroxyproline-rich glycoprotein family protein |
-0.179 |
-0.024 |
| 217 |
259213_at |
AT3G09010
|
[Protein kinase superfamily protein] |
-0.179 |
0.162 |