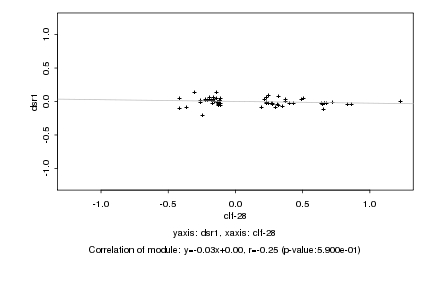
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
clf-28 |
Log2 signal ratio
dsr1 |
| 1 |
254595_at |
AT4G18960
|
AG, AGAMOUS |
1.225 |
0.006 |
| 2 |
248496_at |
AT5G50790
|
AtSWEET10, SWEET10 |
0.857 |
-0.041 |
| 3 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.833 |
-0.038 |
| 4 |
246279_at |
AT4G36740
|
ATHB40, homeobox protein 40, HB-5, HB40, homeobox protein 40 |
0.717 |
-0.007 |
| 5 |
256109_at |
AT1G16950
|
unknown |
0.670 |
-0.022 |
| 6 |
247666_at |
AT5G60140
|
[AP2/B3-like transcriptional factor family protein] |
0.661 |
-0.023 |
| 7 |
246978_at |
AT5G24910
|
CYP714A1, cytochrome P450, family 714, subfamily A, polypeptide 1, ELA1, EUI-like p450 A1 |
0.658 |
-0.026 |
| 8 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.654 |
-0.117 |
| 9 |
264830_at |
AT1G03710
|
[Cystatin/monellin superfamily protein] |
0.646 |
-0.031 |
| 10 |
249497_at |
AT5G39220
|
[alpha/beta-Hydrolases superfamily protein] |
0.640 |
-0.020 |
| 11 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
0.506 |
0.060 |
| 12 |
267129_at |
AT2G23380
|
CLF, CURLY LEAF, ICU1, INCURVATA 1, SDG1, SETDOMAIN GROUP 1, SET1, SETDOMAIN 1 |
0.490 |
0.039 |
| 13 |
256597_at |
AT3G28500
|
[60S acidic ribosomal protein family] |
0.426 |
-0.015 |
| 14 |
263988_at |
AT2G42830
|
AGL5, AGAMOUS-like 5, SHP2, SHATTERPROOF 2 |
0.396 |
-0.017 |
| 15 |
245204_at |
AT5G12270
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.372 |
0.008 |
| 16 |
249939_at |
AT5G22430
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.370 |
0.038 |
| 17 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.348 |
-0.064 |
| 18 |
248015_at |
AT5G56370
|
[F-box/RNI-like/FBD-like domains-containing protein] |
0.320 |
0.089 |
| 19 |
263258_at |
AT1G10540
|
ATNAT8, NAT8, nucleobase-ascorbate transporter 8 |
0.318 |
-0.055 |
| 20 |
260433_at |
AT1G68170
|
UMAMIT23, Usually multiple acids move in and out Transporters 23 |
0.308 |
-0.036 |
| 21 |
255768_at |
AT1G16705
|
[p300/CBP acetyltransferase-related protein-related] |
0.297 |
-0.075 |
| 22 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
0.274 |
-0.038 |
| 23 |
247224_at |
AT5G65080
|
AGL68, AGAMOUS-like 68, MAF5, MADS AFFECTING FLOWERING 5 |
0.273 |
-0.016 |
| 24 |
260841_at |
AT1G29195
|
unknown |
0.261 |
-0.015 |
| 25 |
252896_at |
AT4G39480
|
CYP96A9, cytochrome P450, family 96, subfamily A, polypeptide 9 |
0.245 |
0.099 |
| 26 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.243 |
-0.020 |
| 27 |
251197_at |
AT3G62960
|
[Thioredoxin superfamily protein] |
0.228 |
-0.011 |
| 28 |
264763_at |
AT1G61450
|
unknown |
0.224 |
-0.028 |
| 29 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
0.224 |
0.065 |
| 30 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
0.215 |
0.042 |
| 31 |
259424_at |
AT1G13830
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.193 |
-0.084 |
| 32 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.418 |
0.046 |
| 33 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.418 |
-0.094 |
| 34 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.366 |
-0.085 |
| 35 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.307 |
0.139 |
| 36 |
257035_at |
AT3G19270
|
CYP707A4, cytochrome P450, family 707, subfamily A, polypeptide 4 |
-0.265 |
0.025 |
| 37 |
267535_at |
AT2G41940
|
ZFP8, zinc finger protein 8 |
-0.260 |
-0.004 |
| 38 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.247 |
-0.197 |
| 39 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.226 |
0.030 |
| 40 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
-0.223 |
0.033 |
| 41 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
-0.210 |
0.021 |
| 42 |
250980_at |
AT5G03130
|
unknown |
-0.195 |
0.038 |
| 43 |
257017_at |
AT3G19620
|
[Glycosyl hydrolase family protein] |
-0.194 |
0.068 |
| 44 |
255907_at |
AT1G17920
|
HDG12, homeodomain GLABROUS 12 |
-0.179 |
0.019 |
| 45 |
255766_at |
AT1G16750
|
unknown |
-0.173 |
-0.021 |
| 46 |
266623_at |
AT2G35390
|
[Phosphoribosyltransferase family protein] |
-0.170 |
0.074 |
| 47 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
-0.167 |
0.035 |
| 48 |
247041_at |
AT5G67180
|
TOE3, target of early activation tagged (EAT) 3 |
-0.157 |
0.007 |
| 49 |
264057_at |
AT2G28550
|
RAP2.7, related to AP2.7, TOE1, TARGET OF EARLY ACTIVATION TAGGED (EAT) 1 |
-0.147 |
0.058 |
| 50 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-0.142 |
0.145 |
| 51 |
265029_at |
AT1G24625
|
ZFP7, zinc finger protein 7 |
-0.135 |
-0.012 |
| 52 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
-0.133 |
-0.056 |
| 53 |
246429_at |
AT5G17450
|
HIPP21, heavy metal associated isoprenylated plant protein 21 |
-0.127 |
-0.022 |
| 54 |
251826_at |
AT3G55110
|
ABCG18, ATP-binding cassette G18 |
-0.127 |
-0.042 |
| 55 |
251601_at |
AT3G57800
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.126 |
0.022 |
| 56 |
265985_at |
AT2G24220
|
ATPUP5, purine permease 5, PUP5, purine permease 5 |
-0.123 |
0.016 |
| 57 |
257612_at |
AT3G26600
|
ARO4, armadillo repeat only 4 |
-0.122 |
-0.017 |
| 58 |
248942_at |
AT5G45480
|
unknown |
-0.120 |
0.019 |
| 59 |
255295_at |
AT4G04760
|
[Major facilitator superfamily protein] |
-0.118 |
-0.018 |
| 60 |
263773_at |
AT2G21370
|
XK-1, xylulose kinase-1, XK1, XYLULOSE KINASE 1 |
-0.117 |
0.051 |
| 61 |
255115_at |
AT4G08840
|
APUM11, pumilio 11, PUM11, pumilio 11 |
-0.116 |
-0.045 |