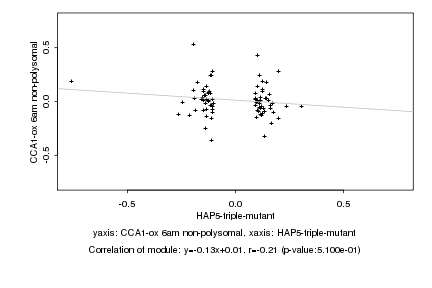
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
HAP5-triple-mutant |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
245712_at |
AT5G04360
|
ATLDA, limit dextrinase, ATPU1, PULLULANASE 1, LDA, limit dextrinase, PU1, PULLULANASE 1 |
0.304 |
-0.037 |
| 2 |
256442_at |
AT3G10930
|
unknown |
0.234 |
-0.041 |
| 3 |
253859_at |
AT4G27657
|
unknown |
0.196 |
-0.150 |
| 4 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.196 |
0.286 |
| 5 |
259001_at |
AT3G01960
|
unknown |
0.173 |
-0.093 |
| 6 |
259850_at |
AT1G72240
|
unknown |
0.169 |
-0.013 |
| 7 |
261684_at |
AT1G47400
|
unknown |
0.165 |
-0.201 |
| 8 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.161 |
-0.031 |
| 9 |
251952_at |
AT3G53650
|
[Histone superfamily protein] |
0.161 |
-0.061 |
| 10 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
0.154 |
0.067 |
| 11 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.152 |
0.017 |
| 12 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.141 |
0.183 |
| 13 |
255535_at |
AT4G01790
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
0.140 |
0.033 |
| 14 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
0.139 |
0.034 |
| 15 |
267077_at |
AT2G40970
|
MYBC1 |
0.132 |
-0.086 |
| 16 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.131 |
-0.319 |
| 17 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
0.127 |
-0.058 |
| 18 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.124 |
-0.104 |
| 19 |
259526_at |
AT1G12570
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
0.121 |
0.193 |
| 20 |
247035_at |
AT5G67110
|
ALC, ALCATRAZ |
0.121 |
0.116 |
| 21 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.121 |
0.096 |
| 22 |
256569_at |
AT3G19550
|
unknown |
0.120 |
-0.125 |
| 23 |
251991_at |
AT3G53340
|
NF-YB10, nuclear factor Y, subunit B10 |
0.119 |
-0.042 |
| 24 |
267141_at |
AT2G38090
|
[Duplicated homeodomain-like superfamily protein] |
0.115 |
0.014 |
| 25 |
253494_at |
AT4G31830
|
unknown |
0.115 |
-0.117 |
| 26 |
250335_at |
AT5G11650
|
[alpha/beta-Hydrolases superfamily protein] |
0.114 |
0.038 |
| 27 |
263985_at |
AT2G42750
|
DJC77, DNA J protein C77 |
0.114 |
-0.059 |
| 28 |
250277_at |
AT5G12940
|
[Leucine-rich repeat (LRR) family protein] |
0.112 |
-0.057 |
| 29 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
0.111 |
-0.055 |
| 30 |
264289_at |
AT1G61890
|
[MATE efflux family protein] |
0.110 |
0.249 |
| 31 |
266320_at |
AT2G46640
|
TAC1, Tiller Angle Control 1 |
0.110 |
0.008 |
| 32 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
0.110 |
-0.017 |
| 33 |
266626_at |
AT2G35360
|
[ubiquitin family protein] |
0.104 |
-0.091 |
| 34 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
0.102 |
0.144 |
| 35 |
249326_at |
AT5G40840
|
AtRAD21.1, Sister chromatid cohesion 1 (SCC1) protein homolog 2, SYN2 |
0.102 |
-0.075 |
| 36 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
0.101 |
0.433 |
| 37 |
249728_at |
AT5G24390
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
0.100 |
0.012 |
| 38 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
0.095 |
-0.000 |
| 39 |
264800_at |
AT1G08800
|
MyoB1, myosin binding protein 1 |
0.093 |
-0.144 |
| 40 |
265214_at |
AT1G05000
|
AtPFA-DSP1, PFA-DSP1, plant and fungi atypical dual-specificity phosphatase 1 |
0.092 |
0.078 |
| 41 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.091 |
-0.029 |
| 42 |
247707_at |
AT5G59450
|
[GRAS family transcription factor] |
0.090 |
0.021 |
| 43 |
259572_at |
AT1G20400
|
unknown |
0.090 |
0.036 |
| 44 |
264190_at |
AT1G54830
|
NF-YC3, nuclear factor Y, subunit C3 |
-0.762 |
0.195 |
| 45 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
-0.266 |
-0.112 |
| 46 |
264650_at |
AT1G08970
|
HAP5C, HEME ACTIVATED PROTEIN 5C, NF-YC9, nuclear factor Y, subunit C9 |
-0.249 |
0.000 |
| 47 |
248021_at |
AT5G56500
|
Cpn60beta3, chaperonin-60beta3 |
-0.216 |
-0.125 |
| 48 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.196 |
0.537 |
| 49 |
266566_at |
AT2G24040
|
[Low temperature and salt responsive protein family] |
-0.195 |
0.109 |
| 50 |
251223_at |
AT3G62610
|
ATMYB11, myb domain protein 11, MYB11, myb domain protein 11, PFG2, PRODUCTION OF FLAVONOL GLYCOSIDES 2 |
-0.193 |
0.029 |
| 51 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
-0.189 |
-0.081 |
| 52 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
-0.177 |
0.179 |
| 53 |
251358_at |
AT3G61160
|
[Protein kinase superfamily protein] |
-0.160 |
0.027 |
| 54 |
256096_at |
AT1G13650
|
unknown |
-0.157 |
0.043 |
| 55 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
-0.153 |
0.020 |
| 56 |
267349_at |
AT2G40010
|
[Ribosomal protein L10 family protein] |
-0.152 |
-0.076 |
| 57 |
245000_at |
ATCG00210
|
YCF6 |
-0.151 |
0.098 |
| 58 |
259527_at |
AT1G12600
|
[UDP-N-acetylglucosamine (UAA) transporter family] |
-0.149 |
0.017 |
| 59 |
262031_x_at |
AT1G37160
|
[gypsy-like retrotransposon family (Athila), similar to putative Athila retroelement ORF1 protein GI:4567296 from (Arabidopsis thaliana)] |
-0.148 |
0.112 |
| 60 |
253796_at |
AT4G28460
|
unknown |
-0.146 |
0.056 |
| 61 |
244942_at |
ATMG00050
|
ORF131 |
-0.143 |
-0.250 |
| 62 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.143 |
-0.017 |
| 63 |
261619_x_at |
AT1G33130
|
[CACTA-like transposase family (Ptta/En/Spm), has a 1.3e-125 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.141 |
0.059 |
| 64 |
246963_at |
AT5G24820
|
[Eukaryotic aspartyl protease family protein] |
-0.137 |
0.025 |
| 65 |
245008_at |
ATCG00360
|
YCF3 |
-0.136 |
0.143 |
| 66 |
253720_at |
AT4G29270
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.136 |
-0.132 |
| 67 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
-0.135 |
-0.073 |
| 68 |
254130_at |
AT4G24540
|
AGL24, AGAMOUS-like 24 |
-0.129 |
0.004 |
| 69 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
-0.129 |
0.075 |
| 70 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
-0.129 |
0.086 |
| 71 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
-0.126 |
0.018 |
| 72 |
245251_at |
AT4G17615
|
ATCBL1, ARABIDOPSIS THALIANA CALCINEURIN B-LIKE PROTEIN, CBL1, calcineurin B-like protein 1, SCABP5, SOS3-LIKE CALCIUM BINDING PROTEIN 5 |
-0.123 |
0.096 |
| 73 |
263281_at |
AT2G14160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.120 |
0.248 |
| 74 |
265128_at |
AT1G30860
|
[RING/U-box superfamily protein] |
-0.117 |
-0.028 |
| 75 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
-0.116 |
0.075 |
| 76 |
260186_at |
AT1G36020
|
unknown |
-0.115 |
-0.154 |
| 77 |
265022_at |
AT1G24520
|
BCP1, homolog of Brassica campestris pollen protein 1 |
-0.112 |
0.248 |
| 78 |
262807_at |
AT1G11740
|
[ankyrin repeat family protein] |
-0.112 |
-0.355 |
| 79 |
261735_at |
AT1G47765
|
[F-box and associated interaction domains-containing protein] |
-0.111 |
-0.034 |
| 80 |
248484_at |
AT5G51030
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.110 |
0.024 |
| 81 |
255391_at |
AT4G03670
|
unknown |
-0.110 |
0.281 |
| 82 |
256099_at |
AT1G13710
|
CYP78A5, cytochrome P450, family 78, subfamily A, polypeptide 5, KLU, KLUH |
-0.109 |
-0.100 |
| 83 |
247666_at |
AT5G60140
|
[AP2/B3-like transcriptional factor family protein] |
-0.109 |
-0.065 |
| 84 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
-0.107 |
-0.037 |
| 85 |
262386_at |
AT1G49370
|
[pseudogene] |
-0.104 |
-0.012 |