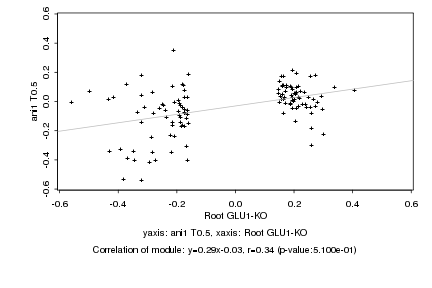
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Root GLU1-KO |
Log2 signal ratio
ani1 T0.5 |
| 1 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
0.404 |
0.077 |
| 2 |
267626_at |
AT2G42250
|
CYP712A1, cytochrome P450, family 712, subfamily A, polypeptide 1 |
0.335 |
0.099 |
| 3 |
263498_at |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
0.299 |
-0.221 |
| 4 |
259991_at |
AT1G68040
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.295 |
-0.054 |
| 5 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
0.293 |
0.040 |
| 6 |
260150_at |
AT1G52820
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.279 |
-0.005 |
| 7 |
247719_at |
AT5G59305
|
unknown |
0.273 |
-0.031 |
| 8 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
0.270 |
0.180 |
| 9 |
260147_at |
AT1G52790
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.264 |
0.014 |
| 10 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.259 |
-0.181 |
| 11 |
249686_at |
AT5G36140
|
CYP716A2, cytochrome P450, family 716, subfamily A, polypeptide 2 |
0.257 |
-0.297 |
| 12 |
259388_at |
AT1G13420
|
ATST4B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 4B, ST4B, sulfotransferase 4B |
0.257 |
-0.076 |
| 13 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.255 |
-0.035 |
| 14 |
250073_at |
AT5G17170
|
ENH1, enhancer of sos3-1 |
0.253 |
0.173 |
| 15 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.249 |
0.030 |
| 16 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
0.241 |
-0.036 |
| 17 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
0.236 |
-0.018 |
| 18 |
252441_at |
AT3G46780
|
PTAC16, plastid transcriptionally active 16 |
0.235 |
0.068 |
| 19 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
0.227 |
-0.015 |
| 20 |
250396_at |
AT5G10970
|
[C2H2 and C2HC zinc fingers superfamily protein] |
0.222 |
0.072 |
| 21 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
0.216 |
0.027 |
| 22 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
0.215 |
0.030 |
| 23 |
249756_at |
AT5G24313
|
unknown |
0.215 |
0.105 |
| 24 |
265923_at |
AT2G18470
|
AtPERK4, proline-rich extensin-like receptor kinase 4, PERK4, proline-rich extensin-like receptor kinase 4 |
0.214 |
-0.033 |
| 25 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.208 |
-0.043 |
| 26 |
264403_at |
AT2G25150
|
[HXXXD-type acyl-transferase family protein] |
0.207 |
0.066 |
| 27 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
0.206 |
0.100 |
| 28 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.205 |
0.193 |
| 29 |
251243_at |
AT3G61870
|
unknown |
0.204 |
0.053 |
| 30 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.204 |
0.060 |
| 31 |
257668_at |
AT3G20460
|
[Major facilitator superfamily protein] |
0.203 |
-0.135 |
| 32 |
266900_at |
AT2G34610
|
unknown |
0.201 |
0.017 |
| 33 |
261533_at |
AT1G71690
|
unknown |
0.201 |
0.057 |
| 34 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
0.198 |
0.007 |
| 35 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
0.194 |
0.213 |
| 36 |
245980_at |
AT5G13140
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.194 |
0.034 |
| 37 |
247577_at |
AT5G61290
|
[Flavin-binding monooxygenase family protein] |
0.193 |
0.008 |
| 38 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
0.192 |
0.084 |
| 39 |
248684_at |
AT5G48485
|
DIR1, DEFECTIVE IN INDUCED RESISTANCE 1 |
0.192 |
-0.047 |
| 40 |
263142_at |
AT1G65230
|
[Uncharacterized conserved protein (DUF2358)] |
0.191 |
0.102 |
| 41 |
256132_at |
AT1G13610
|
[alpha/beta-Hydrolases superfamily protein] |
0.191 |
0.047 |
| 42 |
267569_at |
AT2G30790
|
PSBP-2, photosystem II subunit P-2 |
0.187 |
0.108 |
| 43 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
0.187 |
-0.019 |
| 44 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
0.186 |
-0.008 |
| 45 |
252463_at |
AT3G47070
|
[LOCATED IN: thylakoid, chloroplast thylakoid membrane, chloroplast, chloroplast envelope] |
0.173 |
0.101 |
| 46 |
249694_at |
AT5G35790
|
G6PD1, glucose-6-phosphate dehydrogenase 1 |
0.172 |
0.111 |
| 47 |
251784_at |
AT3G55330
|
PPL1, PsbP-like protein 1 |
0.169 |
-0.008 |
| 48 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
0.169 |
0.071 |
| 49 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
0.165 |
0.028 |
| 50 |
248539_at |
AT5G50130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.163 |
0.020 |
| 51 |
263002_at |
AT1G54200
|
unknown |
0.163 |
-0.076 |
| 52 |
256927_at |
AT3G22550
|
unknown |
0.163 |
0.116 |
| 53 |
266673_at |
AT2G29630
|
PY, PYRIMIDINE REQUIRING, THIC, thiaminC |
0.162 |
0.175 |
| 54 |
256796_at |
AT3G22210
|
unknown |
0.159 |
0.103 |
| 55 |
263761_at |
AT2G21330
|
AtFBA1, FBA1, fructose-bisphosphate aldolase 1 |
0.158 |
0.052 |
| 56 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
0.158 |
0.116 |
| 57 |
251762_at |
AT3G55800
|
SBPASE, sedoheptulose-bisphosphatase |
0.156 |
0.178 |
| 58 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
0.155 |
0.015 |
| 59 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.153 |
0.038 |
| 60 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.149 |
0.000 |
| 61 |
258832_at |
AT3G07070
|
[Protein kinase superfamily protein] |
0.148 |
0.144 |
| 62 |
252327_at |
AT3G48740
|
AtSWEET11, SWEET11 |
0.146 |
0.083 |
| 63 |
249773_at |
AT5G24140
|
SQP2, squalene monooxygenase 2 |
0.144 |
0.061 |
| 64 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
-0.562 |
-0.000 |
| 65 |
255648_at |
AT4G00910
|
[Aluminium activated malate transporter family protein] |
-0.500 |
0.075 |
| 66 |
253502_at |
AT4G31940
|
CYP82C4, cytochrome P450, family 82, subfamily C, polypeptide 4 |
-0.435 |
0.016 |
| 67 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
-0.431 |
-0.342 |
| 68 |
258629_at |
AT3G02850
|
SKOR, STELAR K+ outward rectifier |
-0.419 |
0.033 |
| 69 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
-0.395 |
-0.326 |
| 70 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
-0.385 |
-0.530 |
| 71 |
262795_at |
AT1G13130
|
[Cellulase (glycosyl hydrolase family 5) protein] |
-0.375 |
0.120 |
| 72 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
-0.371 |
-0.389 |
| 73 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
-0.350 |
-0.342 |
| 74 |
245082_at |
AT2G23270
|
unknown |
-0.346 |
-0.402 |
| 75 |
263652_at |
AT1G04330
|
unknown |
-0.336 |
-0.071 |
| 76 |
257135_at |
AT3G12900
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.324 |
0.043 |
| 77 |
250165_at |
AT5G15290
|
CASP5, Casparian strip membrane domain protein 5 |
-0.323 |
0.183 |
| 78 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
-0.322 |
-0.139 |
| 79 |
251755_at |
AT3G55790
|
unknown |
-0.321 |
-0.535 |
| 80 |
264734_at |
AT1G62280
|
SLAH1, SLAC1 homologue 1 |
-0.313 |
-0.036 |
| 81 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
-0.294 |
-0.417 |
| 82 |
259897_at |
AT1G71380
|
ATCEL3, cellulase 3, ATGH9B3, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B3, CEL3, cellulase 3 |
-0.288 |
-0.246 |
| 83 |
264184_at |
AT1G54790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.285 |
0.068 |
| 84 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
-0.285 |
-0.344 |
| 85 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
-0.280 |
-0.077 |
| 86 |
253796_at |
AT4G28460
|
unknown |
-0.276 |
-0.403 |
| 87 |
250455_at |
AT5G09980
|
PROPEP4, elicitor peptide 4 precursor |
-0.261 |
-0.046 |
| 88 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
-0.251 |
-0.017 |
| 89 |
253044_at |
AT4G37290
|
unknown |
-0.248 |
-0.023 |
| 90 |
257757_at |
AT3G18660
|
GUX1, glucuronic acid substitution of xylan 1, PGSIP1, plant glycogenin-like starch initiation protein 1 |
-0.242 |
-0.061 |
| 91 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
-0.236 |
-0.107 |
| 92 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
-0.224 |
-0.232 |
| 93 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
-0.220 |
-0.349 |
| 94 |
257117_at |
AT3G20160
|
[Terpenoid synthases superfamily protein] |
-0.218 |
-0.141 |
| 95 |
264573_at |
AT1G05310
|
[Pectin lyase-like superfamily protein] |
-0.217 |
0.109 |
| 96 |
264940_at |
AT1G60470
|
AtGolS4, galactinol synthase 4, GolS4, galactinol synthase 4 |
-0.215 |
-0.164 |
| 97 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
-0.212 |
0.354 |
| 98 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
-0.211 |
-0.001 |
| 99 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
-0.210 |
-0.235 |
| 100 |
261249_at |
AT1G05880
|
ARI12, ARIADNE 12, ATARI12, ARABIDOPSIS ARIADNE 12 |
-0.197 |
-0.063 |
| 101 |
266282_at |
AT2G29220
|
LecRK-III.1, L-type lectin receptor kinase III.1 |
-0.195 |
0.009 |
| 102 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.194 |
-0.008 |
| 103 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
-0.191 |
-0.092 |
| 104 |
260296_at |
AT1G63750
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.190 |
-0.142 |
| 105 |
249766_at |
AT5G24070
|
[Peroxidase superfamily protein] |
-0.188 |
-0.103 |
| 106 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
-0.188 |
-0.025 |
| 107 |
260522_x_at |
AT2G41730
|
unknown |
-0.185 |
-0.171 |
| 108 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
-0.184 |
-0.162 |
| 109 |
248790_at |
AT5G47450
|
ATTIP2;3, ARABIDOPSIS THALIANA TONOPLAST INTRINSIC PROTEIN 2;3, DELTA-TIP3, DELTA-TONOPLAST INTRINSIC PROTEIN 3, TIP2;3, tonoplast intrinsic protein 2;3 |
-0.183 |
0.118 |
| 110 |
251293_at |
AT3G61930
|
unknown |
-0.181 |
-0.040 |
| 111 |
250958_at |
AT5G03260
|
LAC11, laccase 11 |
-0.180 |
0.110 |
| 112 |
258912_at |
AT3G06460
|
[GNS1/SUR4 membrane protein family] |
-0.177 |
-0.049 |
| 113 |
260394_at |
AT1G74080
|
ATMYB122, MYB DOMAIN PROTEIN 122, MYB122, myb domain protein 122 |
-0.177 |
-0.168 |
| 114 |
266123_at |
AT2G45180
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.176 |
0.077 |
| 115 |
260973_at |
AT1G53490
|
HEI10, homolog of human HEI10 ( Enhancer of cell Invasion No.10) |
-0.176 |
0.029 |
| 116 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
-0.176 |
-0.073 |
| 117 |
260460_at |
AT1G68230
|
[Reticulon family protein] |
-0.174 |
-0.076 |
| 118 |
261429_at |
AT1G18860
|
ATWRKY61, WRKY DNA-BINDING PROTEIN 61, WRKY61, WRKY DNA-binding protein 61 |
-0.169 |
-0.112 |
| 119 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
-0.169 |
-0.302 |
| 120 |
252691_at |
AT3G44050
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.167 |
0.034 |
| 121 |
245767_at |
AT1G33612
|
[Leucine-rich repeat (LRR) family protein] |
-0.167 |
-0.055 |
| 122 |
255252_at |
AT4G04990
|
unknown |
-0.165 |
-0.403 |
| 123 |
267509_at |
AT2G45660
|
AGL20, AGAMOUS-like 20, ATSOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1, SOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1 |
-0.165 |
-0.086 |
| 124 |
248333_at |
AT5G52390
|
[PAR1 protein] |
-0.163 |
0.189 |
| 125 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.161 |
-0.149 |