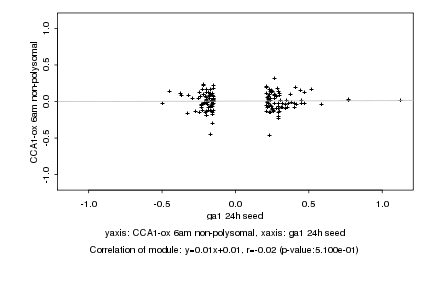
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ga1 24h seed |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
254452_at |
AT4G21100
|
DDB1B, damaged DNA binding protein 1B |
1.122 |
0.014 |
| 2 |
265290_at |
AT2G22590
|
[UDP-Glycosyltransferase superfamily protein] |
0.770 |
0.024 |
| 3 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.764 |
0.029 |
| 4 |
267064_at |
AT2G41110
|
ATCAL5, CAM2, calmodulin 2 |
0.585 |
-0.040 |
| 5 |
267590_at |
AT2G39700
|
ATEXP4, ATEXPA4, expansin A4, ATHEXP ALPHA 1.6, EXPA4, expansin A4 |
0.516 |
0.174 |
| 6 |
251895_at |
AT3G54420
|
ATCHITIV, CHITINASE CLASS IV, ATEP3, homolog of carrot EP3-3 chitinase, CHIV, EP3, homolog of carrot EP3-3 chitinase |
0.470 |
-0.021 |
| 7 |
261819_at |
AT1G11410
|
[S-locus lectin protein kinase family protein] |
0.468 |
0.128 |
| 8 |
251382_at |
AT3G60730
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.448 |
0.016 |
| 9 |
262744_at |
AT1G28680
|
[HXXXD-type acyl-transferase family protein] |
0.447 |
-0.026 |
| 10 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
0.438 |
0.153 |
| 11 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
0.414 |
-0.038 |
| 12 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
0.407 |
0.194 |
| 13 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.399 |
-0.016 |
| 14 |
260392_at |
AT1G74030
|
ENO1, enolase 1 |
0.396 |
-0.075 |
| 15 |
265359_at |
AT2G16720
|
ATMYB7, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 7, ATY49, MYB7, myb domain protein 7 |
0.380 |
-0.002 |
| 16 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.370 |
0.098 |
| 17 |
261129_at |
AT1G04820
|
TOR2, TORTIFOLIA 2, TUA4, tubulin alpha-4 chain |
0.355 |
-0.023 |
| 18 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.353 |
-0.074 |
| 19 |
252024_at |
AT3G52880
|
ATMDAR1, monodehydroascorbate reductase 1, MDAR1, monodehydroascorbate reductase 1 |
0.346 |
0.015 |
| 20 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
0.341 |
-0.092 |
| 21 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.341 |
-0.034 |
| 22 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
0.319 |
-0.017 |
| 23 |
253277_at |
AT4G34230
|
ATCAD5, cinnamyl alcohol dehydrogenase 5, CAD-5, cinnamyl alcohol dehydrogenase 5, CAD5, cinnamyl alcohol dehydrogenase 5 |
0.314 |
-0.080 |
| 24 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
0.313 |
-0.069 |
| 25 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
0.305 |
0.022 |
| 26 |
258962_at |
AT3G10570
|
CYP77A6, cytochrome P450, family 77, subfamily A, polypeptide 6 |
0.296 |
0.123 |
| 27 |
253498_at |
AT4G31890
|
[ARM repeat superfamily protein] |
0.294 |
-0.127 |
| 28 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
0.294 |
0.092 |
| 29 |
265740_at |
AT2G01150
|
RHA2B, RING-H2 finger protein 2B |
0.289 |
0.147 |
| 30 |
266089_at |
AT2G38010
|
[Neutral/alkaline non-lysosomal ceramidase] |
0.289 |
-0.025 |
| 31 |
265144_at |
AT1G51170
|
AGC2-3, AGC2 kinase 3, UCN, UNICORN |
0.288 |
-0.060 |
| 32 |
260561_at |
AT2G43580
|
[Chitinase family protein] |
0.288 |
-0.197 |
| 33 |
258033_at |
AT3G21250
|
ABCC8, ATP-binding cassette C8, ATMRP6, ARABIDOPSIS THALIANA MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 6, MRP6, multidrug resistance-associated protein 6 |
0.288 |
-0.150 |
| 34 |
248864_at |
AT5G46760
|
MYC3 |
0.287 |
-0.086 |
| 35 |
245041_at |
AT2G26530
|
AR781 |
0.287 |
-0.226 |
| 36 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
0.283 |
0.179 |
| 37 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.278 |
0.080 |
| 38 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.277 |
-0.083 |
| 39 |
263002_at |
AT1G54200
|
unknown |
0.269 |
0.090 |
| 40 |
259018_at |
AT3G07390
|
AIR12, Auxin-Induced in Root cultures 12 |
0.266 |
-0.020 |
| 41 |
247279_at |
AT5G64310
|
AGP1, arabinogalactan protein 1, ATAGP1 |
0.265 |
0.088 |
| 42 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.264 |
0.328 |
| 43 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.261 |
0.054 |
| 44 |
249266_at |
AT5G41670
|
[6-phosphogluconate dehydrogenase family protein] |
0.260 |
0.100 |
| 45 |
250611_at |
AT5G07200
|
ATGA20OX3, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 3, GA20OX3, gibberellin 20-oxidase 3, YAP169 |
0.256 |
-0.127 |
| 46 |
262237_at |
AT1G48320
|
DHNAT1, DHNA-CoA thioesterase 1 |
0.254 |
-0.115 |
| 47 |
265475_at |
AT2G15620
|
ATHNIR, ARABIDOPSIS THALIANA NITRITE REDUCTASE, NIR, NITRITE REDUCTASE, NIR1, nitrite reductase 1 |
0.246 |
-0.083 |
| 48 |
256186_at |
AT1G51680
|
4CL.1, 4-COUMARATE:COA LIGASE 1, 4CL1, 4-coumarate:CoA ligase 1, AT4CL1, ARABIDOPSIS THALIANA 4-COUMARATE:COA LIGASE 1 |
0.246 |
0.138 |
| 49 |
251109_at |
AT5G01600
|
ATFER1, ARABIDOPSIS THALIANA FERRETIN 1, FER1, ferretin 1 |
0.246 |
-0.096 |
| 50 |
266246_at |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
0.241 |
0.136 |
| 51 |
260267_at |
AT1G68530
|
CER6, ECERIFERUM 6, CUT1, CUTICULAR 1, G2, KCS6, 3-ketoacyl-CoA synthase 6, POP1, POLLEN-PISTIL INCOMPATIBILITY 1 |
0.241 |
0.154 |
| 52 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.240 |
-0.055 |
| 53 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.237 |
0.043 |
| 54 |
267544_at |
AT2G32720
|
ATCB5-B, ARABIDOPSIS CYTOCHROME B5 ISOFORM B, B5 #4, CB5-B, cytochrome B5 isoform B, CYTB5-D |
0.234 |
0.138 |
| 55 |
249188_at |
AT5G42830
|
[HXXXD-type acyl-transferase family protein] |
0.234 |
-0.150 |
| 56 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
0.233 |
-0.045 |
| 57 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.233 |
0.016 |
| 58 |
256433_at |
AT3G10985
|
ATWI-12, ARABIDOPSIS THALIANA WOUND-INDUCED PROTEIN 12, SAG20, senescence associated gene 20, WI12, WOUND-INDUCED PROTEIN 12 |
0.232 |
-0.137 |
| 59 |
261412_at |
AT1G07890
|
APX1, ascorbate peroxidase 1, ATAPX01, ATAPX1, CS1, MEE6, maternal effect embryo arrest 6 |
0.231 |
0.102 |
| 60 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.230 |
-0.060 |
| 61 |
252133_at |
AT3G50900
|
unknown |
0.229 |
0.165 |
| 62 |
266738_at |
AT2G47010
|
unknown |
0.226 |
-0.462 |
| 63 |
251183_at |
AT3G62630
|
unknown |
0.225 |
0.053 |
| 64 |
256525_at |
AT1G66180
|
[Eukaryotic aspartyl protease family protein] |
0.224 |
0.031 |
| 65 |
263838_at |
AT2G36880
|
MAT3, methionine adenosyltransferase 3 |
0.221 |
0.073 |
| 66 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
0.220 |
-0.025 |
| 67 |
261454_at |
AT1G21090
|
[Cupredoxin superfamily protein] |
0.218 |
0.056 |
| 68 |
255471_at |
AT4G03050
|
AOP3 |
0.213 |
-0.073 |
| 69 |
265188_at |
AT1G23800
|
ALDH2B, aldehyde dehydrogenase 2B, ALDH2B7, aldehyde dehydrogenase 2B7 |
0.213 |
0.006 |
| 70 |
263979_at |
AT2G42840
|
PDF1, protodermal factor 1 |
0.212 |
-0.013 |
| 71 |
264988_at |
AT1G27140
|
ATGSTU14, glutathione S-transferase tau 14, GST13, GLUTATHIONE S-TRANSFERASE 13, GSTU14, glutathione S-transferase tau 14 |
0.210 |
0.203 |
| 72 |
257311_at |
AT3G26570
|
ORF02, PHT2;1, phosphate transporter 2;1 |
0.210 |
-0.130 |
| 73 |
248419_at |
AT5G51550
|
EXL3, EXORDIUM like 3 |
0.208 |
0.209 |
| 74 |
252905_at |
AT4G39720
|
[VQ motif-containing protein] |
0.207 |
-0.041 |
| 75 |
255666_at |
AT4G00390
|
[DNA-binding storekeeper protein-related transcriptional regulator] |
0.207 |
0.122 |
| 76 |
259009_at |
AT3G09260
|
BGLU23, LEB, LONG ER BODY, PSR3.1, PYK10 |
-0.498 |
-0.020 |
| 77 |
254403_at |
AT4G21323
|
[Subtilase family protein] |
-0.450 |
0.143 |
| 78 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-0.377 |
0.123 |
| 79 |
260812_at |
AT1G43650
|
UMAMIT22, Usually multiple acids move in and out Transporters 22 |
-0.368 |
0.083 |
| 80 |
254098_at |
AT4G25100
|
ATFSD1, ARABIDOPSIS FE SUPEROXIDE DISMUTASE 1, FSD1, Fe superoxide dismutase 1 |
-0.327 |
-0.159 |
| 81 |
249547_at |
AT5G38160
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.325 |
0.090 |
| 82 |
247928_at |
AT5G57320
|
VLN5, villin 5 |
-0.294 |
0.047 |
| 83 |
257522_at |
AT3G08990
|
[Yippee family putative zinc-binding protein] |
-0.276 |
-0.124 |
| 84 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
-0.258 |
0.044 |
| 85 |
266438_at |
AT2G43180
|
[Phosphoenolpyruvate carboxylase family protein] |
-0.251 |
0.135 |
| 86 |
260546_at |
AT2G43520
|
ATTI2, trypsin inhibitor protein 2, TI2, trypsin inhibitor protein 2 |
-0.247 |
-0.150 |
| 87 |
252820_at |
AT3G42640
|
AHA8, H(+)-ATPase 8, HA8, H(+)-ATPase 8 |
-0.244 |
0.072 |
| 88 |
248329_at |
AT5G52780
|
unknown |
-0.236 |
-0.039 |
| 89 |
251823_at |
AT3G55080
|
[SET domain-containing protein] |
-0.236 |
-0.050 |
| 90 |
246889_at |
AT5G25470
|
[AP2/B3-like transcriptional factor family protein] |
-0.235 |
-0.074 |
| 91 |
261420_at |
AT1G07720
|
KCS3, 3-ketoacyl-CoA synthase 3 |
-0.228 |
-0.030 |
| 92 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.226 |
0.178 |
| 93 |
247995_at |
AT5G56160
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.225 |
-0.110 |
| 94 |
257421_at |
AT1G12030
|
unknown |
-0.221 |
0.108 |
| 95 |
266300_at |
AT2G01420
|
ATPIN4, ARABIDOPSIS PIN-FORMED 4, PIN4, PIN-FORMED 4 |
-0.220 |
0.229 |
| 96 |
264236_at |
AT1G54680
|
unknown |
-0.218 |
0.237 |
| 97 |
264751_at |
AT1G23020
|
ATFRO3, FERRIC REDUCTION OXIDASE 3, FRO3, ferric reduction oxidase 3 |
-0.217 |
-0.026 |
| 98 |
261540_at |
AT1G63610
|
unknown |
-0.214 |
-0.013 |
| 99 |
261812_at |
AT1G08270
|
unknown |
-0.208 |
0.082 |
| 100 |
252353_at |
AT3G48200
|
unknown |
-0.205 |
-0.141 |
| 101 |
247419_at |
AT5G63080
|
JMJ20, Jumonji domain-containing protein 20 |
-0.203 |
0.057 |
| 102 |
250020_at |
AT5G18180
|
[H/ACA ribonucleoprotein complex, subunit Gar1/Naf1 protein] |
-0.203 |
0.030 |
| 103 |
260669_at |
AT1G19340
|
[Methyltransferase MT-A70 family protein] |
-0.203 |
0.021 |
| 104 |
259252_at |
AT3G07610
|
IBM1, increase in bonsai methylation 1 |
-0.202 |
-0.125 |
| 105 |
260696_at |
AT1G32520
|
unknown |
-0.201 |
0.167 |
| 106 |
247234_at |
AT5G64980
|
unknown |
-0.200 |
0.130 |
| 107 |
257830_at |
AT3G26690
|
ATNUDT13, ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 13, ATNUDX13, nudix hydrolase homolog 13, NUDX13, nudix hydrolase homolog 13 |
-0.199 |
-0.185 |
| 108 |
248959_at |
AT5G45630
|
unknown |
-0.199 |
-0.183 |
| 109 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
-0.198 |
-0.001 |
| 110 |
250773_at |
AT5G05430
|
[RNA-binding protein] |
-0.194 |
0.001 |
| 111 |
251193_at |
AT3G62910
|
APG3, ALBINO AND PALE GREEN, AtcpRF1, Arabidopsis thaliana chloroplast RF1, cpRF1, chloroplast ribosome release factor 1 |
-0.193 |
-0.011 |
| 112 |
259728_at |
AT1G60970
|
[SNARE-like superfamily protein] |
-0.192 |
-0.036 |
| 113 |
251469_at |
AT3G59530
|
LAP3, LESS ADHERENT POLLEN 3 |
-0.189 |
-0.072 |
| 114 |
252506_at |
AT3G46180
|
ATUTR5, UDP-GALACTOSE TRANSPORTER 5, UTR5, UDP-galactose transporter 5 |
-0.188 |
0.137 |
| 115 |
261530_at |
AT1G63460
|
ATGPX8, GPX8, glutathione peroxidase 8 |
-0.187 |
-0.113 |
| 116 |
249900_at |
AT5G22640
|
emb1211, embryo defective 1211, TIC100, TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 100 |
-0.181 |
0.046 |
| 117 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.180 |
0.085 |
| 118 |
251463_at |
AT3G59800
|
unknown |
-0.180 |
0.081 |
| 119 |
265033_at |
AT1G61520
|
LHCA3, photosystem I light harvesting complex gene 3 |
-0.176 |
0.116 |
| 120 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.176 |
0.098 |
| 121 |
255943_at |
AT1G22370
|
AtUGT85A5, UDP-glucosyl transferase 85A5, UGT85A5, UDP-glucosyl transferase 85A5 |
-0.175 |
-0.058 |
| 122 |
267637_at |
AT2G42190
|
unknown |
-0.172 |
-0.447 |
| 123 |
264774_at |
AT1G22890
|
unknown |
-0.171 |
-0.122 |
| 124 |
260323_at |
AT1G63780
|
IMP4 |
-0.170 |
0.178 |
| 125 |
262105_at |
AT1G02810
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.169 |
-0.056 |
| 126 |
245702_at |
AT5G04220
|
ATSYTC, NTMC2T1.3, NTMC2TYPE1.3, SYT3, synaptotagmin 3, SYTC |
-0.167 |
0.021 |
| 127 |
261557_at |
AT1G63640
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
-0.165 |
-0.042 |
| 128 |
252253_at |
AT3G49300
|
[proline-rich family protein] |
-0.164 |
-0.123 |
| 129 |
253597_at |
AT4G30690
|
[Translation initiation factor 3 protein] |
-0.164 |
0.009 |
| 130 |
259520_at |
AT1G12320
|
unknown |
-0.162 |
-0.140 |
| 131 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.162 |
-0.055 |
| 132 |
260220_at |
AT1G74650
|
ATMYB31, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 31, ATY13, MYB31, myb domain protein 31 |
-0.161 |
-0.288 |
| 133 |
264971_at |
AT1G67210
|
[Proline-rich spliceosome-associated (PSP) family protein / zinc knuckle (CCHC-type) family protein] |
-0.161 |
-0.028 |
| 134 |
262646_at |
AT1G62800
|
ASP4, aspartate aminotransferase 4 |
-0.160 |
0.090 |
| 135 |
249966_at |
AT5G19030
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.160 |
-0.172 |
| 136 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
-0.158 |
-0.026 |
| 137 |
246906_at |
AT5G25475
|
[AP2/B3-like transcriptional factor family protein] |
-0.157 |
0.088 |
| 138 |
252593_at |
AT3G45590
|
ATSEN1, splicing endonuclease 1, SEN1, splicing endonuclease 1 |
-0.157 |
0.117 |
| 139 |
257277_at |
AT3G14470
|
[NB-ARC domain-containing disease resistance protein] |
-0.156 |
0.037 |
| 140 |
258880_at |
AT3G06420
|
ATG8H, autophagy 8h |
-0.156 |
0.014 |
| 141 |
263661_at |
AT1G04290
|
[Thioesterase superfamily protein] |
-0.156 |
0.072 |
| 142 |
248401_at |
AT5G52110
|
CCB2, COFACTOR ASSEMBLY OF COMPLEX C, HCF208, HIGH CHLOROPHYLL FLUORESCENCE 208 |
-0.155 |
0.190 |
| 143 |
244908_at |
ATMG00720
|
ORF107D |
-0.155 |
0.008 |
| 144 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.155 |
0.049 |
| 145 |
246303_at |
AT3G51870
|
[Mitochondrial substrate carrier family protein] |
-0.154 |
0.225 |
| 146 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.153 |
0.044 |
| 147 |
250508_at |
AT5G09950
|
MEF7, mitochondrial editing factor 7 |
-0.153 |
-0.017 |
| 148 |
263016_at |
AT1G23410
|
[Ribosomal protein S27a / Ubiquitin family protein] |
-0.152 |
-0.120 |
| 149 |
261800_at |
AT1G30490
|
ATHB9, PHV, PHAVOLUTA |
-0.151 |
0.011 |