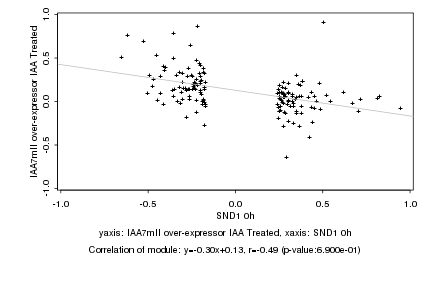
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
SND1 0h |
Log2 signal ratio
IAA7mII over-expressor IAA Treated |
| 1 |
260522_x_at |
AT2G41730
|
unknown |
0.943 |
-0.076 |
| 2 |
245973_at |
AT5G32490
|
[pseudogene] |
0.824 |
0.063 |
| 3 |
251566_at |
AT3G58210
|
[TRAF-like family protein] |
0.809 |
0.044 |
| 4 |
248354_at |
AT5G52330
|
[TRAF-like superfamily protein] |
0.714 |
0.025 |
| 5 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
0.702 |
-0.110 |
| 6 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.670 |
-0.019 |
| 7 |
251366_at |
AT3G61340
|
[F-box and associated interaction domains-containing protein] |
0.616 |
0.112 |
| 8 |
248240_at |
AT5G53950
|
ANAC098, Arabidopsis NAC domain containing protein 98, ATCUC2, CUP-SHAPED COTYLEDON 2, CUC2, CUP-SHAPED COTYLEDON 2 |
0.541 |
0.002 |
| 9 |
245688_at |
AT1G28290
|
AGP31, arabinogalactan protein 31 |
0.520 |
0.073 |
| 10 |
251871_at |
AT3G54520
|
unknown |
0.499 |
0.910 |
| 11 |
247425_at |
AT5G62550
|
unknown |
0.482 |
-0.082 |
| 12 |
254699_at |
AT4G17990
|
unknown |
0.476 |
0.215 |
| 13 |
259409_at |
AT1G13330
|
AHP2, Arabidopsis Hop2 homolog |
0.462 |
0.009 |
| 14 |
267167_at |
AT2G37740
|
ATZFP10, ZINC-FINGER PROTEIN 10, ZFP10, zinc-finger protein 10 |
0.452 |
0.066 |
| 15 |
257386_at |
AT2G42440
|
ASL15, ASYMMETRIC LEAVES 2-like 15, LBD17, LOB domain-containing protein 17 |
0.450 |
-0.074 |
| 16 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
0.436 |
-0.230 |
| 17 |
252215_at |
AT3G50240
|
KICP-02 |
0.433 |
-0.065 |
| 18 |
264892_at |
AT1G23160
|
[Auxin-responsive GH3 family protein] |
0.432 |
0.113 |
| 19 |
257761_at |
AT3G23090
|
WDL3 |
0.420 |
-0.412 |
| 20 |
248241_at |
AT5G53960
|
[ATP binding] |
0.416 |
0.049 |
| 21 |
261869_at |
AT1G11470
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.384 |
0.235 |
| 22 |
265329_at |
AT2G18450
|
SDH1-2, succinate dehydrogenase 1-2 |
0.383 |
0.241 |
| 23 |
259087_at |
AT3G04980
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.376 |
-0.050 |
| 24 |
264720_at |
AT1G70080
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.375 |
-0.134 |
| 25 |
248456_at |
AT5G51380
|
[RNI-like superfamily protein] |
0.374 |
0.062 |
| 26 |
247962_at |
AT5G56580
|
ANQ1, ARABIDOPSIS NQK1, ATMKK6, ARABIDOPSIS THALIANA MAP KINASE KINASE 6, MKK6, MAP kinase kinase 6 |
0.369 |
0.192 |
| 27 |
246016_at |
AT5G10720
|
AHK5, histidine kinase 5, CKI2, CYTOKININ INDEPENDENT 2, HK5, histidine kinase 5 |
0.363 |
-0.284 |
| 28 |
261421_at |
AT1G18840
|
IQD30, IQ-domain 30 |
0.362 |
0.069 |
| 29 |
249060_at |
AT5G44560
|
VPS2.2 |
0.359 |
0.196 |
| 30 |
266687_at |
AT2G19670
|
ATPRMT1A, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 1A, PRMT1A, protein arginine methyltransferase 1A |
0.348 |
-0.114 |
| 31 |
252433_at |
AT3G47560
|
[alpha/beta-Hydrolases superfamily protein] |
0.347 |
0.310 |
| 32 |
264741_at |
AT1G62290
|
[Saposin-like aspartyl protease family protein] |
0.346 |
-0.127 |
| 33 |
248658_at |
AT5G48600
|
ATCAP-C, ARABIDOPSIS THALIANA CHROMOSOME ASSOCIATED PROTEIN-C, ATSMC3, structural maintenance of chromosome 3, ATSMC4, ARABIDOPSIS THALIANA STRUCTURAL MAINTENANCE OF CHROMOSOME 4, SMC3, structural maintenance of chromosome 3 |
0.344 |
0.049 |
| 34 |
247340_at |
AT5G63700
|
[zinc ion binding] |
0.341 |
0.033 |
| 35 |
252430_at |
AT3G47470
|
CAB4, LHCA4, light-harvesting chlorophyll-protein complex I subunit A4 |
0.329 |
-0.252 |
| 36 |
247299_at |
AT5G63900
|
[Acyl-CoA N-acyltransferase with RING/FYVE/PHD-type zinc finger domain] |
0.329 |
-0.056 |
| 37 |
259709_at |
AT1G77655
|
unknown |
0.329 |
-0.021 |
| 38 |
253014_at |
AT4G37940
|
AGL21, AGAMOUS-like 21 |
0.326 |
0.060 |
| 39 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
0.325 |
0.090 |
| 40 |
251761_at |
AT3G55700
|
[UDP-Glycosyltransferase superfamily protein] |
0.322 |
0.005 |
| 41 |
249118_at |
AT5G43870
|
[FUNCTIONS IN: phosphoinositide binding] |
0.311 |
-0.051 |
| 42 |
254341_at |
AT4G22130
|
SRF8, STRUBBELIG-receptor family 8 |
0.303 |
0.211 |
| 43 |
263535_at |
AT2G24970
|
unknown |
0.301 |
0.011 |
| 44 |
262882_at |
AT1G64900
|
CYP89, CYP89A2, cytochrome P450, family 89, subfamily A, polypeptide 2 |
0.300 |
-0.219 |
| 45 |
267529_at |
AT2G45490
|
AtAUR3, ataurora3, AUR3, ataurora3 |
0.300 |
0.099 |
| 46 |
259815_at |
AT1G49870
|
unknown |
0.299 |
0.017 |
| 47 |
247028_at |
AT5G67100
|
ICU2, INCURVATA2 |
0.299 |
0.105 |
| 48 |
248054_at |
AT5G55820
|
WYR, WYRD |
0.298 |
-0.031 |
| 49 |
253791_at |
AT4G28640
|
IAA11, indole-3-acetic acid inducible 11 |
0.289 |
-0.643 |
| 50 |
266087_at |
AT2G37790
|
AKR4C10, Aldo-keto reductase family 4 member C10 |
0.286 |
0.156 |
| 51 |
246585_at |
AT5G14750
|
ATMYB66, myb domain protein 66, MYB66, myb domain protein 66, WER, WEREWOLF, WER1, WEREWOLF 1 |
0.282 |
-0.128 |
| 52 |
250328_at |
AT5G11780
|
unknown |
0.281 |
0.067 |
| 53 |
263360_at |
AT2G03830
|
GLV6, GOLVEN 6, RGF8, root meristem growth factor 8 |
0.279 |
0.089 |
| 54 |
253168_at |
AT4G35070
|
[SBP (S-ribonuclease binding protein) family protein] |
0.278 |
-0.119 |
| 55 |
265422_at |
AT2G20800
|
NDB4, NAD(P)H dehydrogenase B4 |
0.276 |
0.050 |
| 56 |
258341_at |
AT3G22790
|
NET1A, Networked 1A |
0.275 |
0.051 |
| 57 |
248476_at |
AT5G50890
|
[alpha/beta-Hydrolases superfamily protein] |
0.275 |
0.219 |
| 58 |
251373_at |
AT3G60530
|
GATA4, GATA transcription factor 4 |
0.272 |
-0.012 |
| 59 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.272 |
-0.282 |
| 60 |
251239_at |
AT3G62440
|
SAF1, secondary wall thickening-associated F-box 1 |
0.271 |
0.096 |
| 61 |
264966_at |
AT1G60570
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.271 |
0.085 |
| 62 |
259746_at |
AT1G71060
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.270 |
0.090 |
| 63 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
0.267 |
0.100 |
| 64 |
261000_at |
AT1G26540
|
[Agenet domain-containing protein] |
0.265 |
0.170 |
| 65 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.264 |
-0.004 |
| 66 |
264868_at |
AT1G24095
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.261 |
0.021 |
| 67 |
260329_at |
AT1G80370
|
CYCA2;4, Cyclin A2;4 |
0.259 |
0.104 |
| 68 |
252357_at |
AT3G48410
|
[alpha/beta-Hydrolases superfamily protein] |
0.257 |
-0.100 |
| 69 |
249426_at |
AT5G39840
|
[ATP-dependent RNA helicase, mitochondrial, putative] |
0.256 |
0.024 |
| 70 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
0.254 |
-0.048 |
| 71 |
252548_at |
AT3G45850
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.251 |
0.036 |
| 72 |
254821_at |
AT4G12540
|
unknown |
0.251 |
0.067 |
| 73 |
264545_at |
AT1G55670
|
PSAG, photosystem I subunit G |
0.251 |
0.192 |
| 74 |
251898_at |
AT3G54340
|
AP3, APETALA 3, ATAP3 |
0.251 |
0.106 |
| 75 |
266322_at |
AT2G46690
|
SAUR32, SMALL AUXIN UPREGULATED RNA 32 |
0.250 |
-0.111 |
| 76 |
245348_at |
AT4G17770
|
ATTPS5, trehalose phosphatase/synthase 5, TPS5, TREHALOSE -6-PHOSPHATASE SYNTHASE S5, TPS5, trehalose phosphatase/synthase 5 |
0.248 |
-0.114 |
| 77 |
248082_at |
AT5G55400
|
[Actin binding Calponin homology (CH) domain-containing protein] |
0.246 |
0.146 |
| 78 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.245 |
-0.186 |
| 79 |
249276_at |
AT5G41880
|
POLA3, POLA4 |
0.242 |
-0.058 |
| 80 |
257930_at |
AT3G17010
|
REM22, REPRODUCTIVE MERISTEM 22 |
0.240 |
-0.029 |
| 81 |
260565_at |
AT2G43800
|
[Actin-binding FH2 (formin homology 2) family protein] |
0.239 |
0.101 |
| 82 |
247324_at |
AT5G64190
|
unknown |
-0.653 |
0.508 |
| 83 |
246241_at |
AT4G37050
|
AtPLAIVC, phospholipase A IVC, PLA V, PLAIII{beta}, patatin-related phospholipase III beta, PLP4, PATATIN-like protein 4 |
-0.622 |
0.767 |
| 84 |
262356_at |
AT1G73000
|
PYL3, PYR1-like 3, RCAR13, regulatory components of ABA receptor 13 |
-0.529 |
0.701 |
| 85 |
258293_at |
AT3G23430
|
ATPHO1, ARABIDOPSIS PHOSPHATE 1, PHO1, phosphate 1 |
-0.505 |
0.097 |
| 86 |
260048_at |
AT1G73750
|
[Uncharacterised conserved protein UCP031088, alpha/beta hydrolase] |
-0.494 |
0.300 |
| 87 |
254101_at |
AT4G25000
|
AMY1, alpha-amylase-like, ATAMY1 |
-0.476 |
0.174 |
| 88 |
245258_at |
AT4G15340
|
04C11, ATPEN1, pentacyclic triterpene synthase 1, PEN1, pentacyclic triterpene synthase 1 |
-0.471 |
0.262 |
| 89 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
-0.455 |
0.537 |
| 90 |
245044_at |
AT2G26500
|
[cytochrome b6f complex subunit (petM), putative] |
-0.451 |
0.017 |
| 91 |
260806_at |
AT1G78260
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.430 |
0.291 |
| 92 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
-0.430 |
0.094 |
| 93 |
258445_at |
AT3G22400
|
ATLOX5, Arabidopsis thaliana lipoxygenase 5, LOX5 |
-0.416 |
0.409 |
| 94 |
264477_at |
AT1G77240
|
AAE2, acyl activating enzyme 2 |
-0.414 |
-0.024 |
| 95 |
262244_at |
AT1G48260
|
CIPK17, CBL-interacting protein kinase 17, SnRK3.21, SNF1-RELATED PROTEIN KINASE 3.21 |
-0.409 |
0.359 |
| 96 |
252488_at |
AT3G46700
|
[UDP-Glycosyltransferase superfamily protein] |
-0.401 |
0.400 |
| 97 |
261180_at |
AT1G34590
|
unknown |
-0.366 |
0.128 |
| 98 |
254629_at |
AT4G18425
|
unknown |
-0.360 |
0.789 |
| 99 |
245423_at |
AT4G17483
|
[alpha/beta-Hydrolases superfamily protein] |
-0.359 |
0.496 |
| 100 |
262425_at |
AT1G47660
|
unknown |
-0.356 |
0.064 |
| 101 |
259618_at |
AT1G48000
|
AtMYB112, myb domain protein 112, MYB112, myb domain protein 112 |
-0.353 |
0.141 |
| 102 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
-0.342 |
0.303 |
| 103 |
249402_at |
AT5G40155
|
[Defensin-like (DEFL) family protein] |
-0.335 |
0.006 |
| 104 |
248172_at |
AT5G54660
|
[HSP20-like chaperones superfamily protein] |
-0.323 |
0.169 |
| 105 |
246335_at |
AT3G44880
|
ACD1, ACCELERATED CELL DEATH 1, LLS1, LETHAL LEAF-SPOT 1 HOMOLOG, PAO, PHEOPHORBIDE A OXYGENASE |
-0.321 |
0.343 |
| 106 |
251646_at |
AT3G57780
|
unknown |
-0.317 |
-0.020 |
| 107 |
264532_at |
AT1G55740
|
AtSIP1, seed imbibition 1, RS1, raffinose synthase 1, SIP1, seed imbibition 1 |
-0.309 |
0.115 |
| 108 |
255726_at |
AT1G25530
|
[Transmembrane amino acid transporter family protein] |
-0.307 |
0.030 |
| 109 |
265913_at |
AT2G25625
|
unknown |
-0.307 |
0.333 |
| 110 |
251555_at |
AT3G58780
|
AGL1, AGAMOUS-like 1, SHP1, SHATTERPROOF 1 |
-0.304 |
0.229 |
| 111 |
261609_at |
AT1G49740
|
[PLC-like phosphodiesterases superfamily protein] |
-0.299 |
0.155 |
| 112 |
260968_at |
AT1G12250
|
[Pentapeptide repeat-containing protein] |
-0.289 |
0.151 |
| 113 |
261785_at |
AT1G08230
|
[Transmembrane amino acid transporter family protein] |
-0.285 |
0.128 |
| 114 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.283 |
-0.176 |
| 115 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
-0.275 |
0.299 |
| 116 |
246781_at |
AT5G27350
|
SFP1 |
-0.272 |
0.381 |
| 117 |
260582_at |
AT2G47200
|
unknown |
-0.269 |
0.145 |
| 118 |
263668_at |
AT1G04350
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.265 |
0.143 |
| 119 |
260943_at |
AT1G45145
|
ATH5, THIOREDOXIN H-TYPE 5, ATTRX5, thioredoxin H-type 5, LIV1, LOCUS OF INSENSITIVITY TO VICTORIN 1, TRX-h5, THIOREDOXIN H-TYPE 5, TRX5, thioredoxin H-type 5 |
-0.264 |
0.034 |
| 120 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-0.264 |
0.062 |
| 121 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
-0.262 |
0.649 |
| 122 |
257737_at |
AT3G27440
|
UKL5, uridine kinase-like 5 |
-0.253 |
0.304 |
| 123 |
265728_at |
AT2G31990
|
[Exostosin family protein] |
-0.246 |
0.148 |
| 124 |
263258_at |
AT1G10540
|
ATNAT8, NAT8, nucleobase-ascorbate transporter 8 |
-0.246 |
0.295 |
| 125 |
264685_at |
AT1G65610
|
ATGH9A2, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9A2, KOR2, KORRIGAN 2 |
-0.244 |
0.201 |
| 126 |
258323_at |
AT3G22750
|
[Protein kinase superfamily protein] |
-0.242 |
0.178 |
| 127 |
256564_at |
AT3G29770
|
ATMES11, ARABIDOPSIS THALIANA METHYL ESTERASE 11, MES11, methyl esterase 11 |
-0.239 |
0.229 |
| 128 |
260109_at |
AT1G63260
|
TET10, tetraspanin10 |
-0.236 |
0.201 |
| 129 |
253725_at |
AT4G29340
|
PRF4, profilin 4 |
-0.235 |
0.159 |
| 130 |
244949_at |
ATMG00150
|
ORF116 |
-0.232 |
0.217 |
| 131 |
255716_at |
AT4G00330
|
CRCK2, calmodulin-binding receptor-like cytoplasmic kinase 2 |
-0.232 |
0.148 |
| 132 |
257800_at |
AT3G15900
|
unknown |
-0.227 |
0.259 |
| 133 |
245936_at |
AT5G19850
|
[alpha/beta-Hydrolases superfamily protein] |
-0.225 |
-0.116 |
| 134 |
247136_at |
AT5G66170
|
STR18, sulfurtransferase 18 |
-0.224 |
0.021 |
| 135 |
266179_at |
AT2G02300
|
AtPP2-B5, phloem protein 2-B5, PP2-B5, phloem protein 2-B5 |
-0.224 |
0.480 |
| 136 |
246857_at |
AT5G25920
|
unknown |
-0.223 |
0.873 |
| 137 |
266604_at |
AT2G46030
|
UBC6, ubiquitin-conjugating enzyme 6 |
-0.222 |
0.190 |
| 138 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
-0.210 |
0.332 |
| 139 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.210 |
0.442 |
| 140 |
246954_at |
AT5G04830
|
[Nuclear transport factor 2 (NTF2) family protein] |
-0.205 |
0.133 |
| 141 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.205 |
0.292 |
| 142 |
246375_at |
AT1G51830
|
[Leucine-rich repeat protein kinase family protein] |
-0.204 |
0.425 |
| 143 |
251545_at |
AT3G58810
|
ATMTP3, ARABIDOPSIS METAL TOLERANCE PROTEIN 3, ATMTPA2, MTP3, METAL TOLERANCE PROTEIN 3, MTPA2, metal tolerance protein A2 |
-0.204 |
0.233 |
| 144 |
258085_at |
AT3G26100
|
[Regulator of chromosome condensation (RCC1) family protein] |
-0.203 |
0.115 |
| 145 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.202 |
0.218 |
| 146 |
258880_at |
AT3G06420
|
ATG8H, autophagy 8h |
-0.197 |
0.086 |
| 147 |
247734_at |
AT5G59400
|
unknown |
-0.195 |
0.248 |
| 148 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-0.193 |
-0.030 |
| 149 |
256883_at |
AT3G26440
|
unknown |
-0.190 |
0.002 |
| 150 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.187 |
0.021 |
| 151 |
249779_at |
AT5G24230
|
[Lipase class 3-related protein] |
-0.185 |
0.384 |
| 152 |
262183_at |
AT1G77890
|
[DNA-directed RNA polymerase II protein] |
-0.183 |
0.340 |
| 153 |
246197_at |
AT4G37010
|
CEN2, centrin 2 |
-0.183 |
0.029 |
| 154 |
253147_at |
AT4G35600
|
CST, CAST AWAY, CX32, CONNEXIN 32, Kin4, kinase 4 |
-0.182 |
0.139 |
| 155 |
255400_at |
AT4G03760
|
[pseudogene] |
-0.182 |
-0.008 |
| 156 |
255463_at |
AT4G02960
|
ATRE2, retro element 2, RE2, retro element 2 |
-0.180 |
0.171 |
| 157 |
251722_at |
AT3G56200
|
[Transmembrane amino acid transporter family protein] |
-0.179 |
0.333 |
| 158 |
245932_at |
AT5G09290
|
[Inositol monophosphatase family protein] |
-0.178 |
-0.270 |
| 159 |
249807_at |
AT5G23870
|
[Pectinacetylesterase family protein] |
-0.176 |
-0.030 |
| 160 |
261973_at |
AT1G64610
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.175 |
0.230 |
| 161 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
-0.173 |
-0.047 |