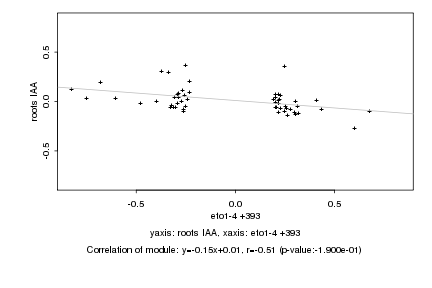
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
eto1-4 +393 |
Log2 signal ratio
roots IAA |
| 1 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.673 |
-0.097 |
| 2 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.599 |
-0.269 |
| 3 |
263070_at |
AT2G17600
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.432 |
-0.076 |
| 4 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
0.404 |
0.014 |
| 5 |
249832_at |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
0.316 |
-0.117 |
| 6 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.312 |
-0.043 |
| 7 |
249134_at |
AT5G43150
|
unknown |
0.302 |
0.010 |
| 8 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
0.302 |
-0.128 |
| 9 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
0.295 |
-0.107 |
| 10 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
0.277 |
-0.071 |
| 11 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
0.260 |
-0.138 |
| 12 |
259308_at |
AT3G05180
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.256 |
-0.066 |
| 13 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
0.248 |
-0.049 |
| 14 |
265083_at |
AT1G03820
|
unknown |
0.244 |
0.361 |
| 15 |
255675_at |
AT4G00480
|
ATMYC1, myc1 |
0.243 |
-0.095 |
| 16 |
265096_at |
AT1G04030
|
unknown |
0.227 |
0.066 |
| 17 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
0.225 |
-0.070 |
| 18 |
251806_at |
AT3G55370
|
OBP3, OBF-binding protein 3 |
0.221 |
0.026 |
| 19 |
267169_at |
AT2G37540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.217 |
-0.105 |
| 20 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
0.217 |
0.077 |
| 21 |
258414_at |
AT3G17380
|
[TRAF-like family protein] |
0.216 |
-0.011 |
| 22 |
260776_at |
AT1G14580
|
[C2H2-like zinc finger protein] |
0.215 |
0.017 |
| 23 |
251553_at |
AT3G58710
|
ATWRKY69, RABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 69, WRKY69, WRKY DNA-binding protein 69 |
0.202 |
-0.051 |
| 24 |
250823_at |
AT5G05180
|
unknown |
0.200 |
0.073 |
| 25 |
261509_at |
AT1G71740
|
unknown |
0.200 |
-0.008 |
| 26 |
266828_at |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
0.199 |
-0.054 |
| 27 |
256754_at |
AT3G25690
|
AtCHUP1, Arabidopsis thaliana CHLOROPLAST UNUSUAL POSITIONING 1, CHUP1, CHLOROPLAST UNUSUAL POSITIONING 1 |
0.197 |
0.048 |
| 28 |
267316_at |
AT2G34710
|
ATHB-14, ATHB14, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 14, PHB, PHABULOSA, PHB-1D, PHABULOSA 1D |
0.189 |
0.024 |
| 29 |
258388_at |
AT3G15370
|
ATEXP12, ATEXPA12, expansin 12, ATHEXP ALPHA 1.24, EXP12, EXPANSIN 12, EXPA12, expansin 12 |
-0.830 |
0.122 |
| 30 |
252571_at |
AT3G45280
|
ATSYP72, SYP72, syntaxin of plants 72 |
-0.755 |
0.034 |
| 31 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
-0.684 |
0.201 |
| 32 |
248509_at |
AT5G50335
|
unknown |
-0.609 |
0.032 |
| 33 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
-0.480 |
-0.012 |
| 34 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
-0.401 |
0.007 |
| 35 |
266908_at |
AT2G34650
|
ABR, ABRUPTUS, PID, PINOID |
-0.374 |
0.309 |
| 36 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
-0.341 |
0.295 |
| 37 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
-0.329 |
-0.059 |
| 38 |
260553_at |
AT2G41800
|
unknown |
-0.324 |
-0.034 |
| 39 |
252740_at |
AT3G43270
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.317 |
-0.051 |
| 40 |
254689_at |
AT4G13710
|
[Pectin lyase-like superfamily protein] |
-0.308 |
0.050 |
| 41 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.306 |
-0.057 |
| 42 |
255039_at |
AT4G09570
|
ATCPK4, CPK4, calcium-dependent protein kinase 4 |
-0.294 |
0.073 |
| 43 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
-0.293 |
-0.015 |
| 44 |
266893_at |
AT2G26070
|
AtRTE1, RTE1, REVERSION-TO-ETHYLENE SENSITIVITY1 |
-0.292 |
0.043 |
| 45 |
267182_at |
AT2G23360
|
unknown |
-0.291 |
0.091 |
| 46 |
258358_at |
AT3G14380
|
[Uncharacterised protein family (UPF0497)] |
-0.272 |
0.007 |
| 47 |
261917_at |
AT1G65920
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
-0.269 |
0.118 |
| 48 |
250204_at |
AT5G13990
|
ATEXO70C2, exocyst subunit exo70 family protein C2, EXO70C2, exocyst subunit exo70 family protein C2 |
-0.266 |
-0.095 |
| 49 |
252888_at |
AT4G39210
|
APL3 |
-0.262 |
-0.079 |
| 50 |
251377_at |
AT3G60650
|
unknown |
-0.259 |
0.063 |
| 51 |
257480_at |
AT1G15640
|
unknown |
-0.255 |
-0.041 |
| 52 |
257026_at |
AT3G19200
|
unknown |
-0.253 |
0.366 |
| 53 |
250201_at |
AT5G14230
|
unknown |
-0.245 |
0.024 |
| 54 |
251946_at |
AT3G53540
|
TRM19, TON1 Recruiting Motif 19 |
-0.233 |
0.206 |
| 55 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.232 |
0.092 |