|
probeID |
AGICode |
Annotation |
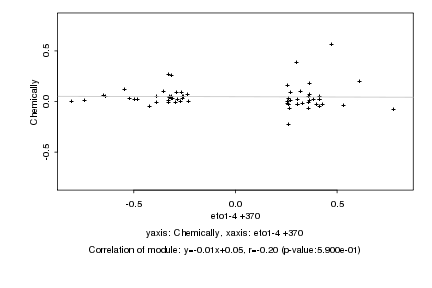
Log2 signal ratio
eto1-4 +370 |
Log2 signal ratio
Chemically |
| 1 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.773 |
-0.077 |
| 2 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
0.606 |
0.206 |
| 3 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.530 |
-0.031 |
| 4 |
251565_at |
AT3G58190
|
ASL16, ASYMMETRIC LEAVES 2-LIKE 16, LBD29, lateral organ boundaries-domain 29 |
0.472 |
0.573 |
| 5 |
246036_at |
AT5G08370
|
AGAL2, alpha-galactosidase 2, AtAGAL2, alpha-galactosidase 2 |
0.426 |
-0.027 |
| 6 |
263070_at |
AT2G17600
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.413 |
0.021 |
| 7 |
250881_at |
AT5G04080
|
unknown |
0.411 |
-0.044 |
| 8 |
261256_at |
AT1G05760
|
RTM1, restricted tev movement 1 |
0.411 |
0.056 |
| 9 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
0.410 |
0.024 |
| 10 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
0.398 |
-0.024 |
| 11 |
266309_at |
AT2G27140
|
[HSP20-like chaperones superfamily protein] |
0.380 |
0.022 |
| 12 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
0.364 |
0.012 |
| 13 |
252184_at |
AT3G50660
|
AtDWF4, CLM, CLOMAZONE-RESISTANT, CYP90B1, CYTOCHROME P450 90B1, DWF4, DWARF 4, PSC1, PARTIALLY SUPPRESSING COI1 INSENSITIVITY TO JA 1, SAV1, SHADE AVOIDANCE 1, SNP2, SUPPRESSOR OF NPH4 2 |
0.363 |
0.078 |
| 14 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
0.360 |
0.183 |
| 15 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
0.357 |
-0.063 |
| 16 |
265277_at |
AT2G28410
|
unknown |
0.357 |
-0.009 |
| 17 |
264697_at |
AT1G70210
|
ATCYCD1;1, CYCD1;1, CYCLIN D1;1 |
0.356 |
0.053 |
| 18 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
0.325 |
-0.019 |
| 19 |
251167_at |
AT3G63360
|
[defensin-related] |
0.316 |
0.101 |
| 20 |
255893_at |
AT1G17960
|
[Threonyl-tRNA synthetase] |
0.305 |
-0.021 |
| 21 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
0.305 |
-0.026 |
| 22 |
249832_at |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
0.303 |
0.021 |
| 23 |
265083_at |
AT1G03820
|
unknown |
0.299 |
0.387 |
| 24 |
245423_at |
AT4G17483
|
[alpha/beta-Hydrolases superfamily protein] |
0.268 |
0.013 |
| 25 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
0.268 |
0.097 |
| 26 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
0.262 |
-0.064 |
| 27 |
249309_at |
AT5G41410
|
BEL1, BELL 1 |
0.260 |
-0.024 |
| 28 |
263932_at |
AT2G35990
|
LOG2, LONELY GUY 2 |
0.257 |
-0.026 |
| 29 |
261509_at |
AT1G71740
|
unknown |
0.257 |
-0.225 |
| 30 |
251806_at |
AT3G55370
|
OBP3, OBF-binding protein 3 |
0.257 |
0.033 |
| 31 |
267169_at |
AT2G37540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.254 |
-0.018 |
| 32 |
266828_at |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
0.252 |
0.009 |
| 33 |
265096_at |
AT1G04030
|
unknown |
0.252 |
0.160 |
| 34 |
252571_at |
AT3G45280
|
ATSYP72, SYP72, syntaxin of plants 72 |
-0.810 |
0.007 |
| 35 |
258388_at |
AT3G15370
|
ATEXP12, ATEXPA12, expansin 12, ATHEXP ALPHA 1.24, EXP12, EXPANSIN 12, EXPA12, expansin 12 |
-0.744 |
0.012 |
| 36 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
-0.652 |
0.067 |
| 37 |
261367_at |
AT1G53080
|
[Legume lectin family protein] |
-0.643 |
0.057 |
| 38 |
250509_at |
AT5G09970
|
CYP78A7, cytochrome P450, family 78, subfamily A, polypeptide 7 |
-0.547 |
0.119 |
| 39 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
-0.525 |
0.034 |
| 40 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
-0.497 |
0.024 |
| 41 |
250049_at |
AT5G17780
|
[alpha/beta-Hydrolases superfamily protein] |
-0.486 |
0.027 |
| 42 |
255814_at |
AT1G19900
|
[glyoxal oxidase-related protein] |
-0.423 |
-0.046 |
| 43 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.390 |
-0.001 |
| 44 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
-0.389 |
0.055 |
| 45 |
252888_at |
AT4G39210
|
APL3 |
-0.358 |
0.103 |
| 46 |
265994_at |
AT2G24170
|
[Endomembrane protein 70 protein family] |
-0.334 |
-0.006 |
| 47 |
247753_at |
AT5G59070
|
[UDP-Glycosyltransferase superfamily protein] |
-0.333 |
0.016 |
| 48 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
-0.331 |
0.275 |
| 49 |
253809_at |
AT4G28320
|
AtMAN5, MAN5, endo-beta-mannase 5 |
-0.329 |
0.047 |
| 50 |
267182_at |
AT2G23360
|
unknown |
-0.325 |
0.051 |
| 51 |
266908_at |
AT2G34650
|
ABR, ABRUPTUS, PID, PINOID |
-0.323 |
0.042 |
| 52 |
255039_at |
AT4G09570
|
ATCPK4, CPK4, calcium-dependent protein kinase 4 |
-0.319 |
0.039 |
| 53 |
252866_at |
AT4G39840
|
unknown |
-0.316 |
0.058 |
| 54 |
254689_at |
AT4G13710
|
[Pectin lyase-like superfamily protein] |
-0.316 |
0.262 |
| 55 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.310 |
0.039 |
| 56 |
248717_at |
AT5G48175
|
unknown |
-0.295 |
-0.000 |
| 57 |
247097_at |
AT5G66460
|
AtMAN7, MAN7, endo-beta-mannase 7 |
-0.294 |
0.091 |
| 58 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
-0.288 |
0.027 |
| 59 |
251373_at |
AT3G60530
|
GATA4, GATA transcription factor 4 |
-0.272 |
0.001 |
| 60 |
261467_at |
AT1G28520
|
ATVOZ1, VASCULAR PLANT ONE ZINC FINGER PROTEIN, VOZ1, vascular plant one zinc finger protein |
-0.266 |
0.091 |
| 61 |
264510_at |
AT1G09530
|
PAP3, PHYTOCHROME-ASSOCIATED PROTEIN 3, PIF3, phytochrome interacting factor 3, POC1, PHOTOCURRENT 1 |
-0.263 |
0.038 |
| 62 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
-0.261 |
0.057 |
| 63 |
258358_at |
AT3G14380
|
[Uncharacterised protein family (UPF0497)] |
-0.257 |
0.050 |
| 64 |
252312_at |
AT3G49380
|
iqd15, IQ-domain 15 |
-0.237 |
0.075 |
| 65 |
245094_at |
AT2G40840
|
DPE2, disproportionating enzyme 2 |
-0.233 |
0.008 |