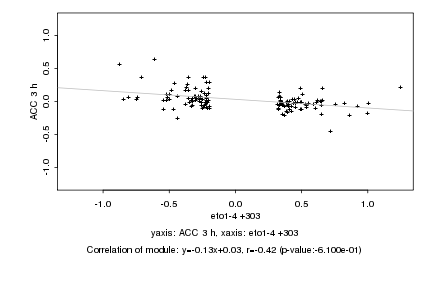
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
eto1-4 +303 |
Log2 signal ratio
ACC 3 h |
| 1 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
1.244 |
0.220 |
| 2 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
1.003 |
-0.027 |
| 3 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.996 |
-0.169 |
| 4 |
259653_at |
AT1G55240
|
unknown |
0.920 |
-0.061 |
| 5 |
256368_at |
AT1G66800
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.860 |
-0.197 |
| 6 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.820 |
-0.015 |
| 7 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
0.749 |
-0.038 |
| 8 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.717 |
-0.445 |
| 9 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.654 |
0.204 |
| 10 |
267485_at |
AT2G02820
|
AtMYB88, myb domain protein 88, MYB88, myb domain protein 88 |
0.652 |
0.028 |
| 11 |
249202_at |
AT5G42580
|
CYP705A12, cytochrome P450, family 705, subfamily A, polypeptide 12 |
0.649 |
-0.188 |
| 12 |
249614_at |
AT5G37300
|
WSD1 |
0.647 |
0.003 |
| 13 |
247135_at |
AT5G66260
|
SAUR11, SMALL AUXIN UPREGULATED RNA 11 |
0.643 |
-0.046 |
| 14 |
246947_at |
AT5G25120
|
CYP71B11, ytochrome p450, family 71, subfamily B, polypeptide 11 |
0.638 |
0.006 |
| 15 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.618 |
0.030 |
| 16 |
263281_at |
AT2G14160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.612 |
-0.014 |
| 17 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.601 |
-0.093 |
| 18 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
0.585 |
-0.036 |
| 19 |
251565_at |
AT3G58190
|
ASL16, ASYMMETRIC LEAVES 2-LIKE 16, LBD29, lateral organ boundaries-domain 29 |
0.545 |
-0.027 |
| 20 |
263070_at |
AT2G17600
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.536 |
-0.048 |
| 21 |
250375_at |
AT5G11540
|
AtGulLO3, GulLO3, L -gulono-1,4-lactone ( L -GulL) oxidase 3 |
0.533 |
-0.078 |
| 22 |
264296_at |
AT1G78720
|
[SecY protein transport family protein] |
0.522 |
-0.031 |
| 23 |
264379_at |
AT2G25200
|
unknown |
0.501 |
0.115 |
| 24 |
254761_at |
AT4G13195
|
CLE44, CLAVATA3/ESR-RELATED 44 |
0.497 |
-0.116 |
| 25 |
251768_at |
AT3G55940
|
[Phosphoinositide-specific phospholipase C family protein] |
0.494 |
-0.014 |
| 26 |
253024_at |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
0.492 |
-0.000 |
| 27 |
263607_at |
AT2G16270
|
unknown |
0.488 |
-0.009 |
| 28 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
0.485 |
0.201 |
| 29 |
251358_at |
AT3G61160
|
[Protein kinase superfamily protein] |
0.484 |
-0.117 |
| 30 |
250881_at |
AT5G04080
|
unknown |
0.473 |
0.047 |
| 31 |
245880_at |
AT5G09430
|
[alpha/beta-Hydrolases superfamily protein] |
0.464 |
-0.005 |
| 32 |
246036_at |
AT5G08370
|
AGAL2, alpha-galactosidase 2, AtAGAL2, alpha-galactosidase 2 |
0.452 |
-0.083 |
| 33 |
265024_at |
AT1G24600
|
unknown |
0.449 |
-0.020 |
| 34 |
261256_at |
AT1G05760
|
RTM1, restricted tev movement 1 |
0.448 |
-0.001 |
| 35 |
252458_at |
AT3G47210
|
unknown |
0.440 |
0.034 |
| 36 |
246072_at |
AT5G20240
|
PI, PISTILLATA |
0.434 |
-0.016 |
| 37 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
0.428 |
0.041 |
| 38 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.418 |
-0.146 |
| 39 |
266309_at |
AT2G27140
|
[HSP20-like chaperones superfamily protein] |
0.416 |
-0.068 |
| 40 |
264697_at |
AT1G70210
|
ATCYCD1;1, CYCD1;1, CYCLIN D1;1 |
0.404 |
0.009 |
| 41 |
253851_at |
AT4G28110
|
AtMYB41, myb domain protein 41, MYB41, myb domain protein 41 |
0.401 |
-0.112 |
| 42 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.397 |
-0.031 |
| 43 |
262264_at |
AT1G42470
|
[Patched family protein] |
0.396 |
-0.069 |
| 44 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
0.395 |
-0.042 |
| 45 |
266708_at |
AT2G03200
|
[Eukaryotic aspartyl protease family protein] |
0.393 |
-0.057 |
| 46 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
0.388 |
0.009 |
| 47 |
253827_at |
AT4G28085
|
unknown |
0.387 |
-0.157 |
| 48 |
252184_at |
AT3G50660
|
AtDWF4, CLM, CLOMAZONE-RESISTANT, CYP90B1, CYTOCHROME P450 90B1, DWF4, DWARF 4, PSC1, PARTIALLY SUPPRESSING COI1 INSENSITIVITY TO JA 1, SAV1, SHADE AVOIDANCE 1, SNP2, SUPPRESSOR OF NPH4 2 |
0.381 |
-0.142 |
| 49 |
252195_at |
AT3G50190
|
unknown |
0.379 |
-0.009 |
| 50 |
265277_at |
AT2G28410
|
unknown |
0.368 |
-0.210 |
| 51 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.366 |
-0.062 |
| 52 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
0.356 |
-0.049 |
| 53 |
254032_at |
AT4G25940
|
[ENTH/ANTH/VHS superfamily protein] |
0.355 |
-0.051 |
| 54 |
254142_at |
AT4G24630
|
[DHHC-type zinc finger family protein] |
0.353 |
-0.009 |
| 55 |
249309_at |
AT5G41410
|
BEL1, BELL 1 |
0.352 |
-0.190 |
| 56 |
266984_at |
AT2G39570
|
ACR9, ACT domain repeats 9 |
0.346 |
-0.012 |
| 57 |
249134_at |
AT5G43150
|
unknown |
0.342 |
-0.029 |
| 58 |
263407_at |
AT2G04038
|
AtbZIP48, basic leucine-zipper 48, bZIP48, basic leucine-zipper 48 |
0.339 |
0.008 |
| 59 |
260011_at |
AT1G68110
|
[ENTH/ANTH/VHS superfamily protein] |
0.339 |
0.067 |
| 60 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
0.338 |
0.065 |
| 61 |
252484_at |
AT3G46690
|
[UDP-Glycosyltransferase superfamily protein] |
0.335 |
0.030 |
| 62 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
0.334 |
-0.040 |
| 63 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.331 |
0.145 |
| 64 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.330 |
0.084 |
| 65 |
245174_at |
AT2G47500
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
0.329 |
-0.057 |
| 66 |
250223_at |
AT5G14070
|
ROXY2 |
0.328 |
-0.040 |
| 67 |
262120_at |
AT1G02950
|
ATGSTF4, glutathione S-transferase F4, GST31, GLUTATHIONE S-TRANSFERASE 31, GSTF4, glutathione S-transferase F4 |
0.325 |
0.066 |
| 68 |
262072_at |
AT1G59590
|
ZCF37 |
0.324 |
-0.115 |
| 69 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
0.318 |
-0.091 |
| 70 |
257896_at |
AT3G16920
|
ATCTL2, CTL2, chitinase-like protein 2 |
0.316 |
-0.039 |
| 71 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
-0.880 |
0.573 |
| 72 |
258388_at |
AT3G15370
|
ATEXP12, ATEXPA12, expansin 12, ATHEXP ALPHA 1.24, EXP12, EXPANSIN 12, EXPA12, expansin 12 |
-0.850 |
0.038 |
| 73 |
252571_at |
AT3G45280
|
ATSYP72, SYP72, syntaxin of plants 72 |
-0.812 |
0.063 |
| 74 |
248048_at |
AT5G56080
|
ATNAS2, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 2, NAS2, nicotianamine synthase 2 |
-0.752 |
0.040 |
| 75 |
256602_at |
AT3G28310
|
unknown |
-0.745 |
0.076 |
| 76 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.712 |
0.377 |
| 77 |
246000_at |
AT5G20820
|
SAUR76, SMALL AUXIN UPREGULATED RNA 76 |
-0.619 |
0.641 |
| 78 |
261367_at |
AT1G53080
|
[Legume lectin family protein] |
-0.551 |
-0.110 |
| 79 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
-0.545 |
0.028 |
| 80 |
256600_at |
AT3G14850
|
TBL41, TRICHOME BIREFRINGENCE-LIKE 41 |
-0.526 |
0.109 |
| 81 |
254754_at |
AT4G13210
|
[Pectin lyase-like superfamily protein] |
-0.524 |
0.016 |
| 82 |
249010_at |
AT5G44580
|
unknown |
-0.518 |
0.064 |
| 83 |
258241_at |
AT3G27650
|
LBD25, LOB domain-containing protein 25 |
-0.505 |
0.116 |
| 84 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
-0.502 |
0.041 |
| 85 |
250049_at |
AT5G17780
|
[alpha/beta-Hydrolases superfamily protein] |
-0.485 |
0.178 |
| 86 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-0.468 |
-0.106 |
| 87 |
252103_at |
AT3G51410
|
unknown |
-0.466 |
0.284 |
| 88 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.445 |
-0.246 |
| 89 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.439 |
0.085 |
| 90 |
261368_at |
AT1G53070
|
[Legume lectin family protein] |
-0.381 |
-0.040 |
| 91 |
248623_at |
AT5G49170
|
unknown |
-0.380 |
0.173 |
| 92 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
-0.371 |
0.214 |
| 93 |
250509_at |
AT5G09970
|
CYP78A7, cytochrome P450, family 78, subfamily A, polypeptide 7 |
-0.367 |
0.266 |
| 94 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
-0.359 |
0.375 |
| 95 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
-0.359 |
0.048 |
| 96 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.358 |
0.173 |
| 97 |
261084_at |
AT1G07440
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.348 |
-0.013 |
| 98 |
251952_at |
AT3G53650
|
[Histone superfamily protein] |
-0.337 |
0.020 |
| 99 |
264565_at |
AT1G05280
|
unknown |
-0.334 |
-0.064 |
| 100 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.328 |
0.050 |
| 101 |
260553_at |
AT2G41800
|
unknown |
-0.327 |
-0.057 |
| 102 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
-0.327 |
0.010 |
| 103 |
252118_at |
AT3G51400
|
unknown |
-0.315 |
0.080 |
| 104 |
254689_at |
AT4G13710
|
[Pectin lyase-like superfamily protein] |
-0.309 |
0.030 |
| 105 |
261470_at |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
-0.309 |
0.203 |
| 106 |
247049_at |
AT5G66440
|
unknown |
-0.302 |
0.082 |
| 107 |
261917_at |
AT1G65920
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
-0.299 |
0.051 |
| 108 |
257026_at |
AT3G19200
|
unknown |
-0.283 |
0.088 |
| 109 |
252312_at |
AT3G49380
|
iqd15, IQ-domain 15 |
-0.277 |
0.015 |
| 110 |
258782_at |
AT3G11750
|
FOLB1 |
-0.274 |
0.040 |
| 111 |
256360_at |
AT1G66440
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.270 |
0.058 |
| 112 |
255039_at |
AT4G09570
|
ATCPK4, CPK4, calcium-dependent protein kinase 4 |
-0.268 |
0.085 |
| 113 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.261 |
-0.059 |
| 114 |
264510_at |
AT1G09530
|
PAP3, PHYTOCHROME-ASSOCIATED PROTEIN 3, PIF3, phytochrome interacting factor 3, POC1, PHOTOCURRENT 1 |
-0.260 |
0.154 |
| 115 |
258358_at |
AT3G14380
|
[Uncharacterised protein family (UPF0497)] |
-0.259 |
-0.003 |
| 116 |
252343_at |
AT3G48610
|
NPC6, non-specific phospholipase C6 |
-0.256 |
0.044 |
| 117 |
255055_at |
AT4G09810
|
[Nucleotide-sugar transporter family protein] |
-0.250 |
-0.098 |
| 118 |
256721_at |
AT2G34150
|
ATRANGAP2, ATSCAR1, SCAR1, WAVE1, WISKOTT-ALDRICH SYNDROME PROTEIN FAMILY VERPROLIN HOMOLOGOUS PROTEIN 1 |
-0.246 |
-0.080 |
| 119 |
253155_at |
AT4G35720
|
unknown |
-0.243 |
0.367 |
| 120 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
-0.240 |
0.122 |
| 121 |
258629_at |
AT3G02850
|
SKOR, STELAR K+ outward rectifier |
-0.239 |
-0.060 |
| 122 |
257357_at |
AT2G41050
|
[PQ-loop repeat family protein / transmembrane family protein] |
-0.233 |
-0.042 |
| 123 |
245143_at |
AT2G45450
|
ZPR1, LITTLE ZIPPER 1 |
-0.229 |
-0.013 |
| 124 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.228 |
0.369 |
| 125 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
-0.226 |
0.297 |
| 126 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
-0.225 |
0.046 |
| 127 |
246815_at |
AT5G27220
|
[Frigida-like protein] |
-0.223 |
0.063 |
| 128 |
249167_at |
AT5G42860
|
unknown |
-0.220 |
-0.088 |
| 129 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
-0.220 |
0.103 |
| 130 |
248172_at |
AT5G54660
|
[HSP20-like chaperones superfamily protein] |
-0.219 |
0.027 |
| 131 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.219 |
-0.013 |
| 132 |
259743_at |
AT1G71140
|
[MATE efflux family protein] |
-0.217 |
-0.012 |
| 133 |
265945_at |
AT2G19500
|
ATCKX2, CKX2, cytokinin oxidase 2 |
-0.212 |
-0.102 |
| 134 |
266119_at |
AT2G02100
|
LCR69, low-molecular-weight cysteine-rich 69, PDF2.2 |
-0.211 |
0.024 |
| 135 |
254318_at |
AT4G22530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.208 |
0.123 |
| 136 |
251244_at |
AT3G62060
|
[Pectinacetylesterase family protein] |
-0.204 |
0.207 |
| 137 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
-0.201 |
-0.071 |
| 138 |
257875_at |
AT3G17120
|
unknown |
-0.196 |
-0.099 |
| 139 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
-0.196 |
0.299 |