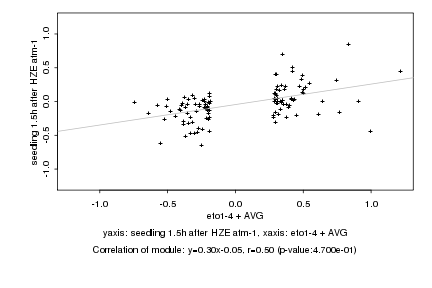
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
eto1-4 + AVG |
Log2 signal ratio
seedling 1.5h after HZE atm-1 |
| 1 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
1.214 |
0.449 |
| 2 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
0.991 |
-0.436 |
| 3 |
259653_at |
AT1G55240
|
unknown |
0.904 |
0.006 |
| 4 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.830 |
0.848 |
| 5 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.765 |
-0.152 |
| 6 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
0.741 |
0.319 |
| 7 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.640 |
0.012 |
| 8 |
267485_at |
AT2G02820
|
AtMYB88, myb domain protein 88, MYB88, myb domain protein 88 |
0.609 |
-0.183 |
| 9 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.541 |
0.278 |
| 10 |
250881_at |
AT5G04080
|
unknown |
0.510 |
0.221 |
| 11 |
260460_at |
AT1G68230
|
[Reticulon family protein] |
0.497 |
0.178 |
| 12 |
266309_at |
AT2G27140
|
[HSP20-like chaperones superfamily protein] |
0.495 |
0.125 |
| 13 |
261256_at |
AT1G05760
|
RTM1, restricted tev movement 1 |
0.492 |
0.146 |
| 14 |
260011_at |
AT1G68110
|
[ENTH/ANTH/VHS superfamily protein] |
0.491 |
0.399 |
| 15 |
251358_at |
AT3G61160
|
[Protein kinase superfamily protein] |
0.481 |
0.329 |
| 16 |
247710_at |
AT5G59260
|
LecRK-II.1, L-type lectin receptor kinase II.1 |
0.468 |
0.224 |
| 17 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
0.444 |
-0.195 |
| 18 |
247116_at |
AT5G65970
|
ATMLO10, MILDEW RESISTANCE LOCUS O 10, MLO10, MILDEW RESISTANCE LOCUS O 10 |
0.433 |
0.036 |
| 19 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
0.424 |
0.019 |
| 20 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.416 |
0.515 |
| 21 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
0.415 |
0.453 |
| 22 |
264697_at |
AT1G70210
|
ATCYCD1;1, CYCD1;1, CYCLIN D1;1 |
0.413 |
0.050 |
| 23 |
265024_at |
AT1G24600
|
unknown |
0.411 |
0.033 |
| 24 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
0.392 |
-0.050 |
| 25 |
266708_at |
AT2G03200
|
[Eukaryotic aspartyl protease family protein] |
0.389 |
-0.080 |
| 26 |
265277_at |
AT2G28410
|
unknown |
0.374 |
-0.232 |
| 27 |
261090_at |
AT1G07560
|
[Leucine-rich repeat protein kinase family protein] |
0.371 |
-0.039 |
| 28 |
254142_at |
AT4G24630
|
[DHHC-type zinc finger family protein] |
0.366 |
0.226 |
| 29 |
251577_at |
AT3G58350
|
RTM3, RESTRICTED TEV MOVEMENT 3 |
0.359 |
0.187 |
| 30 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.347 |
-0.041 |
| 31 |
264529_at |
AT1G30820
|
[CTP synthase family protein] |
0.346 |
0.017 |
| 32 |
259982_at |
AT1G76410
|
ATL8 |
0.344 |
0.697 |
| 33 |
257896_at |
AT3G16920
|
ATCTL2, CTL2, chitinase-like protein 2 |
0.342 |
0.025 |
| 34 |
265188_at |
AT1G23800
|
ALDH2B, aldehyde dehydrogenase 2B, ALDH2B7, aldehyde dehydrogenase 2B7 |
0.339 |
-0.002 |
| 35 |
262264_at |
AT1G42470
|
[Patched family protein] |
0.338 |
0.239 |
| 36 |
258414_at |
AT3G17380
|
[TRAF-like family protein] |
0.326 |
0.010 |
| 37 |
249309_at |
AT5G41410
|
BEL1, BELL 1 |
0.325 |
-0.116 |
| 38 |
266984_at |
AT2G39570
|
ACR9, ACT domain repeats 9 |
0.320 |
0.173 |
| 39 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
0.312 |
-0.178 |
| 40 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
0.310 |
0.227 |
| 41 |
259022_at |
AT3G07420
|
ATNS2, ASPARAGINYL-TRNA SYNTHETASE 2, NS2, asparaginyl-tRNA synthetase 2, SYNC2, SYNTHETASE C2, SYNC2_ARATH, ASPARAGINYL-TRNA SYNTHETASE 2 |
0.306 |
0.059 |
| 42 |
264933_at |
AT1G61160
|
unknown |
0.305 |
0.010 |
| 43 |
255528_at |
AT4G02090
|
unknown |
0.303 |
-0.027 |
| 44 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.302 |
0.409 |
| 45 |
249202_at |
AT5G42580
|
CYP705A12, cytochrome P450, family 705, subfamily A, polypeptide 12 |
0.300 |
0.093 |
| 46 |
251009_at |
AT5G02640
|
unknown |
0.299 |
-0.028 |
| 47 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
0.297 |
0.192 |
| 48 |
259149_at |
AT3G10340
|
PAL4, phenylalanine ammonia-lyase 4 |
0.293 |
-0.161 |
| 49 |
245174_at |
AT2G47500
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
0.293 |
-0.299 |
| 50 |
253454_at |
AT4G31875
|
unknown |
0.291 |
0.408 |
| 51 |
248855_at |
AT5G46590
|
anac096, NAC domain containing protein 96, NAC096, NAC domain containing protein 96 |
0.289 |
0.115 |
| 52 |
265096_at |
AT1G04030
|
unknown |
0.288 |
0.129 |
| 53 |
258203_at |
AT3G13950
|
unknown |
0.288 |
0.042 |
| 54 |
262072_at |
AT1G59590
|
ZCF37 |
0.287 |
0.130 |
| 55 |
257535_at |
AT3G09490
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.286 |
0.059 |
| 56 |
250475_at |
AT5G10180
|
AST68, ARABIDOPSIS SULFATE TRANSPORTER 68, SULTR2;1, sulfate transporter 2;1 |
0.286 |
0.009 |
| 57 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
0.278 |
-0.236 |
| 58 |
266956_at |
AT2G34510
|
unknown |
0.277 |
-0.194 |
| 59 |
254459_at |
AT4G21200
|
ATGA2OX8, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 8, GA2OX8, gibberellin 2-oxidase 8 |
-0.749 |
-0.013 |
| 60 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.645 |
-0.167 |
| 61 |
261691_at |
AT1G50060
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.581 |
-0.051 |
| 62 |
252661_at |
AT3G44450
|
unknown |
-0.557 |
-0.619 |
| 63 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
-0.526 |
-0.266 |
| 64 |
250467_at |
AT5G10100
|
TPPI, trehalose-6-phosphate phosphatase I |
-0.509 |
-0.073 |
| 65 |
257081_at |
AT3G30460
|
[RING/U-box superfamily protein] |
-0.508 |
0.031 |
| 66 |
249010_at |
AT5G44580
|
unknown |
-0.484 |
-0.140 |
| 67 |
245676_at |
AT1G56670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.444 |
-0.209 |
| 68 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.413 |
-0.118 |
| 69 |
248873_at |
AT5G46450
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.407 |
-0.125 |
| 70 |
264767_at |
AT1G61380
|
SD1-29, S-domain-1 29 |
-0.399 |
-0.046 |
| 71 |
262819_at |
AT1G11600
|
CYP77B1, cytochrome P450, family 77, subfamily B, polypeptide 1 |
-0.395 |
-0.020 |
| 72 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.390 |
-0.293 |
| 73 |
252343_at |
AT3G48610
|
NPC6, non-specific phospholipase C6 |
-0.387 |
-0.331 |
| 74 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
-0.377 |
0.065 |
| 75 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.375 |
-0.080 |
| 76 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.372 |
-0.509 |
| 77 |
264565_at |
AT1G05280
|
unknown |
-0.359 |
-0.169 |
| 78 |
246016_at |
AT5G10720
|
AHK5, histidine kinase 5, CKI2, CYTOKININ INDEPENDENT 2, HK5, histidine kinase 5 |
-0.357 |
-0.034 |
| 79 |
261368_at |
AT1G53070
|
[Legume lectin family protein] |
-0.351 |
-0.324 |
| 80 |
257026_at |
AT3G19200
|
unknown |
-0.347 |
0.034 |
| 81 |
250479_at |
AT5G10260
|
AtRABH1e, RAB GTPase homolog H1E, RABH1e, RAB GTPase homolog H1E |
-0.334 |
-0.228 |
| 82 |
257533_at |
AT3G10840
|
[alpha/beta-Hydrolases superfamily protein] |
-0.332 |
-0.464 |
| 83 |
262605_at |
AT1G15170
|
[MATE efflux family protein] |
-0.321 |
0.095 |
| 84 |
252888_at |
AT4G39210
|
APL3 |
-0.318 |
-0.308 |
| 85 |
251377_at |
AT3G60650
|
unknown |
-0.307 |
0.052 |
| 86 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.305 |
-0.469 |
| 87 |
261084_at |
AT1G07440
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.301 |
-0.042 |
| 88 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
-0.293 |
-0.138 |
| 89 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
-0.282 |
-0.454 |
| 90 |
254043_at |
AT4G25990
|
CIL |
-0.279 |
-0.393 |
| 91 |
245094_at |
AT2G40840
|
DPE2, disproportionating enzyme 2 |
-0.267 |
-0.033 |
| 92 |
255055_at |
AT4G09810
|
[Nucleotide-sugar transporter family protein] |
-0.265 |
-0.062 |
| 93 |
264745_at |
AT1G62180
|
5'adenylylphosphosulfate reductase 2 |
-0.257 |
-0.649 |
| 94 |
254982_at |
AT4G10470
|
unknown |
-0.247 |
0.024 |
| 95 |
250413_at |
AT5G11160
|
APT5, adenine phosphoribosyltransferase 5 |
-0.244 |
-0.401 |
| 96 |
266124_at |
AT2G45080
|
cycp3;1, cyclin p3;1 |
-0.230 |
0.041 |
| 97 |
256360_at |
AT1G66440
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.229 |
0.007 |
| 98 |
263199_at |
AT1G05590
|
ATHEX3, BETA-HEXOSAMINIDASE 3, HEXO2, beta-hexosaminidase 2 |
-0.229 |
-0.077 |
| 99 |
248091_at |
AT5G55120
|
galactose-1-phosphate guanylyltransferase (GDP)s;GDP-D-glucose phosphorylases;quercetin 4'-O-glucosyltransferases |
-0.224 |
-0.100 |
| 100 |
266950_at |
AT2G18910
|
[hydroxyproline-rich glycoprotein family protein] |
-0.223 |
-0.032 |
| 101 |
263330_at |
AT2G15320
|
[Leucine-rich repeat (LRR) family protein] |
-0.219 |
-0.241 |
| 102 |
258929_at |
AT3G10060
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.211 |
-0.060 |
| 103 |
250073_at |
AT5G17170
|
ENH1, enhancer of sos3-1 |
-0.211 |
-0.074 |
| 104 |
265283_at |
AT2G20370
|
AtMUR3, KAM1, KATAMARI 1, MUR3, MURUS 3, RSA3, SHORT ROOT IN SALT MEDIUM 3 |
-0.208 |
-0.104 |
| 105 |
266591_at |
AT2G46225
|
ABIL1, ABI-1-like 1 |
-0.207 |
-0.127 |
| 106 |
248006_at |
AT5G56250
|
HAP8, HAPLESS 8 |
-0.205 |
0.000 |
| 107 |
266464_at |
AT2G47800
|
ABCC4, ATP-binding cassette C4, ATMRP4, multidrug resistance-associated protein 4, EST3, MRP4, multidrug resistance-associated protein 4 |
-0.203 |
-0.254 |
| 108 |
264340_at |
AT1G70280
|
[NHL domain-containing protein] |
-0.200 |
-0.165 |
| 109 |
246516_at |
AT5G15740
|
[O-fucosyltransferase family protein] |
-0.198 |
-0.431 |
| 110 |
251255_at |
AT3G62280
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.197 |
0.119 |
| 111 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
-0.196 |
0.087 |
| 112 |
266131_at |
AT2G45160
|
ATHAM1, ARABIDOPSIS THALIANA HAIRY MERISTEM 1, HAM1, HAIRY MERISTEM 1, LOM1, LOST MERISTEMS 1 |
-0.196 |
-0.016 |
| 113 |
267055_at |
AT2G38360
|
PRA1.B4, prenylated RAB acceptor 1.B4 |
-0.194 |
-0.234 |
| 114 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-0.193 |
-0.124 |
| 115 |
263003_at |
AT1G54450
|
[Calcium-binding EF-hand family protein] |
-0.187 |
0.008 |