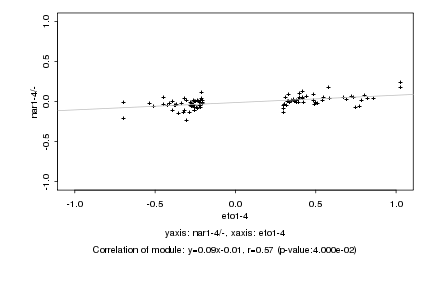
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
eto1-4 |
Log2 signal ratio
nar1-4/- |
| 1 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
1.025 |
0.240 |
| 2 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
1.022 |
0.179 |
| 3 |
258388_at |
AT3G15370
|
ATEXP12, ATEXPA12, expansin 12, ATHEXP ALPHA 1.24, EXP12, EXPANSIN 12, EXPA12, expansin 12 |
0.857 |
0.043 |
| 4 |
252571_at |
AT3G45280
|
ATSYP72, SYP72, syntaxin of plants 72 |
0.816 |
0.038 |
| 5 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
0.799 |
0.082 |
| 6 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
0.784 |
0.015 |
| 7 |
256602_at |
AT3G28310
|
unknown |
0.771 |
-0.056 |
| 8 |
248509_at |
AT5G50335
|
unknown |
0.744 |
-0.074 |
| 9 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
0.730 |
0.057 |
| 10 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.721 |
0.073 |
| 11 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
0.688 |
0.027 |
| 12 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.669 |
0.052 |
| 13 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.584 |
0.038 |
| 14 |
263194_at |
AT1G36060
|
[Integrase-type DNA-binding superfamily protein] |
0.579 |
0.182 |
| 15 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.543 |
0.057 |
| 16 |
261367_at |
AT1G53080
|
[Legume lectin family protein] |
0.538 |
0.015 |
| 17 |
250049_at |
AT5G17780
|
[alpha/beta-Hydrolases superfamily protein] |
0.510 |
-0.022 |
| 18 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
0.493 |
-0.006 |
| 19 |
258241_at |
AT3G27650
|
LBD25, LOB domain-containing protein 25 |
0.486 |
-0.025 |
| 20 |
250509_at |
AT5G09970
|
CYP78A7, cytochrome P450, family 78, subfamily A, polypeptide 7 |
0.485 |
0.016 |
| 21 |
253054_at |
AT4G37580
|
COP3, CONSTITUTIVE PHOTOMORPHOGENIC 3, HLS1, HOOKLESS 1, UNS2, UNUSUAL SUGAR RESPONSE 2 |
0.480 |
0.088 |
| 22 |
257026_at |
AT3G19200
|
unknown |
0.441 |
0.067 |
| 23 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
0.420 |
-0.010 |
| 24 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
0.416 |
0.134 |
| 25 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
0.415 |
0.055 |
| 26 |
246935_at |
AT5G25350
|
EBF2, EIN3-binding F box protein 2 |
0.412 |
0.048 |
| 27 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
0.394 |
0.101 |
| 28 |
249087_at |
AT5G44210
|
ATERF-9, ERF DOMAIN PROTEIN- 9, ATERF9, ERF DOMAIN PROTEIN 9, ERF9, erf domain protein 9 |
0.394 |
0.056 |
| 29 |
266908_at |
AT2G34650
|
ABR, ABRUPTUS, PID, PINOID |
0.392 |
-0.001 |
| 30 |
262981_at |
AT1G75590
|
SAUR52, SMALL AUXIN UPREGULATED RNA 52 |
0.388 |
0.026 |
| 31 |
261470_at |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
0.379 |
-0.010 |
| 32 |
251373_at |
AT3G60530
|
GATA4, GATA transcription factor 4 |
0.376 |
0.022 |
| 33 |
247020_at |
AT5G67020
|
unknown |
0.366 |
0.010 |
| 34 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
0.359 |
0.037 |
| 35 |
247753_at |
AT5G59070
|
[UDP-Glycosyltransferase superfamily protein] |
0.347 |
0.023 |
| 36 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.333 |
-0.004 |
| 37 |
263680_at |
AT1G26930
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.331 |
-0.007 |
| 38 |
249010_at |
AT5G44580
|
unknown |
0.329 |
0.094 |
| 39 |
251946_at |
AT3G53540
|
TRM19, TON1 Recruiting Motif 19 |
0.327 |
0.012 |
| 40 |
245098_at |
AT2G40940
|
ERS, ETHYLENE RESPONSE SENSOR, ERS1, ethylene response sensor 1 |
0.325 |
0.011 |
| 41 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.318 |
0.001 |
| 42 |
251377_at |
AT3G60650
|
unknown |
0.317 |
-0.038 |
| 43 |
258029_at |
AT3G27580
|
ATPK7, D6PKL3, D6 PROTEIN KINASE LIKE 3 |
0.307 |
0.053 |
| 44 |
259966_at |
AT1G76500
|
AHL29, AT-hook motif nuclear-localized protein 29, SOB3, SUPPRESSOR OF PHYB-4#3 |
0.302 |
-0.033 |
| 45 |
248777_at |
AT5G47920
|
unknown |
0.301 |
-0.029 |
| 46 |
260639_at |
AT1G53180
|
unknown |
0.299 |
-0.075 |
| 47 |
250204_at |
AT5G13990
|
ATEXO70C2, exocyst subunit exo70 family protein C2, EXO70C2, exocyst subunit exo70 family protein C2 |
0.299 |
-0.125 |
| 48 |
256137_at |
AT1G48690
|
[Auxin-responsive GH3 family protein] |
0.295 |
-0.038 |
| 49 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-0.701 |
0.000 |
| 50 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
-0.697 |
-0.205 |
| 51 |
267485_at |
AT2G02820
|
AtMYB88, myb domain protein 88, MYB88, myb domain protein 88 |
-0.535 |
-0.014 |
| 52 |
251768_at |
AT3G55940
|
[Phosphoinositide-specific phospholipase C family protein] |
-0.511 |
-0.054 |
| 53 |
247135_at |
AT5G66260
|
SAUR11, SMALL AUXIN UPREGULATED RNA 11 |
-0.454 |
-0.028 |
| 54 |
249832_at |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
-0.449 |
0.054 |
| 55 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.425 |
-0.042 |
| 56 |
250375_at |
AT5G11540
|
AtGulLO3, GulLO3, L -gulono-1,4-lactone ( L -GulL) oxidase 3 |
-0.415 |
-0.024 |
| 57 |
265543_at |
AT2G28270
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.395 |
-0.104 |
| 58 |
256754_at |
AT3G25690
|
AtCHUP1, Arabidopsis thaliana CHLOROPLAST UNUSUAL POSITIONING 1, CHUP1, CHLOROPLAST UNUSUAL POSITIONING 1 |
-0.392 |
0.001 |
| 59 |
261256_at |
AT1G05760
|
RTM1, restricted tev movement 1 |
-0.382 |
-0.046 |
| 60 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
-0.378 |
-0.049 |
| 61 |
256966_at |
AT3G13400
|
sks13, SKU5 similar 13 |
-0.370 |
-0.032 |
| 62 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.360 |
-0.146 |
| 63 |
249614_at |
AT5G37300
|
WSD1 |
-0.337 |
-0.018 |
| 64 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.328 |
-0.127 |
| 65 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
-0.319 |
0.040 |
| 66 |
265463_at |
AT2G37090
|
IRX9, IRREGULAR XYLEM 9 |
-0.319 |
-0.106 |
| 67 |
265277_at |
AT2G28410
|
unknown |
-0.308 |
0.025 |
| 68 |
261090_at |
AT1G07560
|
[Leucine-rich repeat protein kinase family protein] |
-0.307 |
-0.229 |
| 69 |
264697_at |
AT1G70210
|
ATCYCD1;1, CYCD1;1, CYCLIN D1;1 |
-0.287 |
-0.129 |
| 70 |
259985_at |
AT1G76620
|
unknown |
-0.285 |
-0.007 |
| 71 |
245546_at |
AT4G15290
|
ATCSLB05, ATCSLB5, CELLULOSE SYNTHASE LIKE 5, CSLB05 |
-0.280 |
-0.046 |
| 72 |
265096_at |
AT1G04030
|
unknown |
-0.274 |
-0.014 |
| 73 |
256626_at |
AT3G20015
|
[Eukaryotic aspartyl protease family protein] |
-0.274 |
-0.056 |
| 74 |
249288_at |
AT5G41050
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.267 |
0.017 |
| 75 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
-0.266 |
-0.030 |
| 76 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
-0.265 |
-0.048 |
| 77 |
254150_at |
AT4G24350
|
[Phosphorylase superfamily protein] |
-0.259 |
-0.067 |
| 78 |
257896_at |
AT3G16920
|
ATCTL2, CTL2, chitinase-like protein 2 |
-0.258 |
-0.101 |
| 79 |
266956_at |
AT2G34510
|
unknown |
-0.258 |
0.007 |
| 80 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.257 |
-0.105 |
| 81 |
263238_at |
AT2G16580
|
SAUR8, SMALL AUXIN UPREGULATED RNA 8 |
-0.255 |
-0.068 |
| 82 |
260143_at |
AT1G71880
|
ATSUC1, ARABIDOPSIS THALIANA SUCROSE-PROTON SYMPORTER 1, SUC1, sucrose-proton symporter 1 |
-0.252 |
0.011 |
| 83 |
259022_at |
AT3G07420
|
ATNS2, ASPARAGINYL-TRNA SYNTHETASE 2, NS2, asparaginyl-tRNA synthetase 2, SYNC2, SYNTHETASE C2, SYNC2_ARATH, ASPARAGINYL-TRNA SYNTHETASE 2 |
-0.248 |
-0.069 |
| 84 |
260776_at |
AT1G14580
|
[C2H2-like zinc finger protein] |
-0.240 |
0.014 |
| 85 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
-0.239 |
-0.076 |
| 86 |
258414_at |
AT3G17380
|
[TRAF-like family protein] |
-0.236 |
0.007 |
| 87 |
247853_at |
AT5G58140
|
NPL1, NON PHOTOTROPIC HYPOCOTYL 1-LIKE, PHOT2, phototropin 2 |
-0.227 |
-0.005 |
| 88 |
264745_at |
AT1G62180
|
5'adenylylphosphosulfate reductase 2 |
-0.226 |
-0.049 |
| 89 |
253278_at |
AT4G34220
|
[Leucine-rich repeat protein kinase family protein] |
-0.223 |
0.016 |
| 90 |
265305_at |
AT2G20340
|
AAS, aromatic aldehyde synthase, AtAAS |
-0.221 |
-0.067 |
| 91 |
257535_at |
AT3G09490
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.218 |
-0.040 |
| 92 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
-0.216 |
0.123 |
| 93 |
264626_at |
AT1G65620
|
AS2, ASYMMETRIC LEAVES 2 |
-0.212 |
0.039 |
| 94 |
267137_at |
AT2G23410
|
ACPT, cis-prenyltransferase, AtcPT3, CPT, cis-prenyltransferase, cPT3, cis-prenyltransferase 3 |
-0.211 |
-0.016 |
| 95 |
255410_at |
AT4G03100
|
[Rho GTPase activating protein with PAK-box/P21-Rho-binding domain] |
-0.211 |
0.022 |
| 96 |
258183_at |
AT3G21550
|
AtDMP2, Arabidopsis thaliana DUF679 domain membrane protein 2, DMP2, DUF679 domain membrane protein 2 |
-0.207 |
-0.012 |