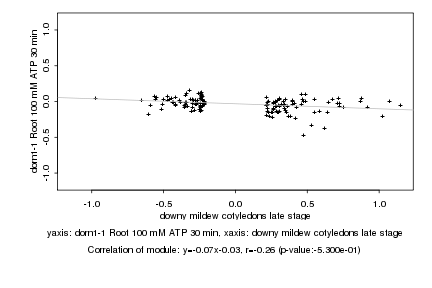
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
downy mildew cotyledons late stage |
Log2 signal ratio
dorn1-1 Root 100 mM ATP 30 min |
| 1 |
259385_at |
AT1G13470
|
unknown |
1.146 |
-0.048 |
| 2 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
1.072 |
0.008 |
| 3 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
1.022 |
-0.203 |
| 4 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
0.913 |
-0.080 |
| 5 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
0.873 |
0.052 |
| 6 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
0.867 |
0.013 |
| 7 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.752 |
-0.070 |
| 8 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.724 |
-0.014 |
| 9 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
0.718 |
-0.063 |
| 10 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
0.711 |
0.048 |
| 11 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.707 |
-0.027 |
| 12 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
0.670 |
0.030 |
| 13 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
0.647 |
-0.013 |
| 14 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.635 |
-0.143 |
| 15 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.616 |
-0.375 |
| 16 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.584 |
-0.129 |
| 17 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
0.548 |
0.036 |
| 18 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
0.546 |
-0.142 |
| 19 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.524 |
-0.323 |
| 20 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.486 |
0.104 |
| 21 |
263032_at |
AT1G23850
|
unknown |
0.485 |
0.007 |
| 22 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.473 |
-0.464 |
| 23 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
0.473 |
0.011 |
| 24 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
0.460 |
0.030 |
| 25 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
0.459 |
0.106 |
| 26 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.454 |
-0.031 |
| 27 |
258787_at |
AT3G11840
|
PUB24, plant U-box 24 |
0.421 |
-0.079 |
| 28 |
253405_at |
AT4G32800
|
[Integrase-type DNA-binding superfamily protein] |
0.412 |
-0.234 |
| 29 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
0.408 |
-0.026 |
| 30 |
261585_at |
AT1G01010
|
ANAC001, NAC domain containing protein 1, NAC001, NAC domain containing protein 1 |
0.397 |
-0.037 |
| 31 |
251633_at |
AT3G57460
|
[catalytics] |
0.393 |
0.024 |
| 32 |
247835_at |
AT5G57910
|
unknown |
0.392 |
0.011 |
| 33 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.379 |
-0.206 |
| 34 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.363 |
-0.205 |
| 35 |
259518_at |
AT1G20510
|
OPCL1, OPC-8:0 CoA ligase1 |
0.362 |
-0.064 |
| 36 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.353 |
-0.151 |
| 37 |
262542_at |
AT1G34180
|
anac016, NAC domain containing protein 16, NAC016, NAC domain containing protein 16 |
0.348 |
-0.121 |
| 38 |
253401_at |
AT4G32870
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.344 |
0.031 |
| 39 |
256593_at |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.339 |
-0.085 |
| 40 |
251259_at |
AT3G62260
|
[Protein phosphatase 2C family protein] |
0.337 |
-0.030 |
| 41 |
255406_at |
AT4G03450
|
[Ankyrin repeat family protein] |
0.331 |
0.008 |
| 42 |
258982_at |
AT3G08870
|
LecRK-VI.1, L-type lectin receptor kinase VI.1 |
0.331 |
-0.043 |
| 43 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.314 |
-0.039 |
| 44 |
254275_at |
AT4G22670
|
AtHip1, HSP70-interacting protein 1, HIP1, HSP70-interacting protein 1, TPR11, tetratricopeptide repeat 11 |
0.306 |
0.035 |
| 45 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
0.304 |
0.043 |
| 46 |
255502_at |
AT4G02410
|
AtLPK1, LecRK-IV.3, L-type lectin receptor kinase IV.3, LPK1, lectin-like protein kinase 1 |
0.304 |
-0.127 |
| 47 |
258507_at |
AT3G06500
|
A/N-InvC, alkaline/neutral invertase C |
0.301 |
-0.061 |
| 48 |
250858_at |
AT5G04760
|
[Duplicated homeodomain-like superfamily protein] |
0.297 |
-0.050 |
| 49 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.296 |
-0.143 |
| 50 |
264636_at |
AT1G65490
|
unknown |
0.289 |
-0.144 |
| 51 |
254318_at |
AT4G22530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.287 |
0.025 |
| 52 |
266993_at |
AT2G39210
|
[Major facilitator superfamily protein] |
0.280 |
-0.139 |
| 53 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.280 |
-0.061 |
| 54 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
0.277 |
-0.059 |
| 55 |
266977_at |
AT2G39420
|
[alpha/beta-Hydrolases superfamily protein] |
0.277 |
0.004 |
| 56 |
255787_at |
AT2G33590
|
AtCRL1, CRL1, CCR(Cinnamoyl coA:NADP oxidoreductase)-like 1 |
0.275 |
-0.056 |
| 57 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.273 |
-0.001 |
| 58 |
254784_at |
AT4G12720
|
AtNUDT7, Arabidopsis thaliana Nudix hydrolase homolog 7, GFG1, GROWTH FACTOR GENE 1, NUDT7 |
0.264 |
-0.105 |
| 59 |
247351_at |
AT5G63790
|
ANAC102, NAC domain containing protein 102, NAC102, NAC domain containing protein 102 |
0.262 |
-0.014 |
| 60 |
246631_at |
AT1G50740
|
[Transmembrane proteins 14C] |
0.260 |
-0.090 |
| 61 |
254673_at |
AT4G18430
|
AtRABA1e, RAB GTPase homolog A1E, RABA1e, RAB GTPase homolog A1E |
0.258 |
-0.001 |
| 62 |
259418_at |
AT1G02390
|
ATGPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2, GPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2 |
0.255 |
-0.216 |
| 63 |
249777_at |
AT5G24210
|
[alpha/beta-Hydrolases superfamily protein] |
0.251 |
-0.144 |
| 64 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
0.249 |
-0.147 |
| 65 |
265728_at |
AT2G31990
|
[Exostosin family protein] |
0.231 |
-0.196 |
| 66 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
0.226 |
0.012 |
| 67 |
254573_at |
AT4G19420
|
[Pectinacetylesterase family protein] |
0.224 |
-0.143 |
| 68 |
253284_at |
AT4G34150
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.222 |
-0.130 |
| 69 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.220 |
-0.009 |
| 70 |
267490_at |
AT2G19130
|
[S-locus lectin protein kinase family protein] |
0.219 |
-0.035 |
| 71 |
256529_at |
AT1G33260
|
[Protein kinase superfamily protein] |
0.217 |
-0.092 |
| 72 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
0.217 |
-0.041 |
| 73 |
245731_at |
AT1G73500
|
ATMKK9, MKK9, MAP kinase kinase 9 |
0.214 |
-0.049 |
| 74 |
262254_at |
AT1G53920
|
GLIP5, GDSL-motif lipase 5 |
0.212 |
0.067 |
| 75 |
248959_at |
AT5G45630
|
unknown |
0.212 |
-0.050 |
| 76 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.211 |
-0.185 |
| 77 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
-0.979 |
0.048 |
| 78 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
-0.658 |
0.019 |
| 79 |
256602_at |
AT3G28310
|
unknown |
-0.611 |
-0.179 |
| 80 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.592 |
-0.048 |
| 81 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
-0.564 |
0.081 |
| 82 |
256631_at |
AT3G28320
|
unknown |
-0.560 |
0.033 |
| 83 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.556 |
0.057 |
| 84 |
253322_at |
AT4G33980
|
unknown |
-0.521 |
-0.108 |
| 85 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.508 |
-0.037 |
| 86 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
-0.506 |
0.030 |
| 87 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
-0.479 |
0.017 |
| 88 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
-0.478 |
0.075 |
| 89 |
259595_at |
AT1G28050
|
BBX13, B-box domain protein 13 |
-0.462 |
0.042 |
| 90 |
250194_at |
AT5G14550
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.451 |
0.056 |
| 91 |
246525_at |
AT5G15840
|
BBX1, B-box domain protein 1, CO, CONSTANS, FG |
-0.440 |
-0.006 |
| 92 |
267280_at |
AT2G19450
|
ABX45, AS11, ATDGAT, Arabidopsis thaliana acyl-CoA:diacylglycerol acyltransferase, AtDGAT1, DGAT1, acyl-CoA:diacylglycerol acyltransferase 1, RDS1, TAG1, TRIACYLGLYCEROL BIOSYNTHESIS DEFECT 1 |
-0.438 |
-0.009 |
| 93 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.422 |
-0.040 |
| 94 |
247525_at |
AT5G61380
|
APRR1, AtTOC1, TIMING OF CAB EXPRESSION 1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TOC1, TIMING OF CAB EXPRESSION 1 |
-0.422 |
-0.050 |
| 95 |
251753_at |
AT3G55760
|
unknown |
-0.421 |
0.062 |
| 96 |
262892_at |
AT1G79440
|
ALDH5F1, aldehyde dehydrogenase 5F1, ENF1, ENLARGED FIL EXPRESSION DOMAIN 1, SSADH, SUCCINIC SEMIALDEHYDE DEHYDROGENASE, SSADH1, SUCCINIC SEMIALDEHYDE DEHYDROGENASE 1 |
-0.395 |
0.016 |
| 97 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
-0.383 |
-0.011 |
| 98 |
266839_at |
AT2G25930
|
ELF3, EARLY FLOWERING 3, PYK20 |
-0.357 |
-0.051 |
| 99 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.356 |
-0.079 |
| 100 |
249053_at |
AT5G44440
|
[FAD-binding Berberine family protein] |
-0.354 |
-0.002 |
| 101 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
-0.352 |
0.097 |
| 102 |
261203_at |
AT1G12845
|
unknown |
-0.351 |
-0.032 |
| 103 |
247216_at |
AT5G64860
|
DPE1, disproportionating enzyme |
-0.349 |
-0.057 |
| 104 |
255070_at |
AT4G09020
|
ATISA3, ISA3, isoamylase 3 |
-0.346 |
-0.001 |
| 105 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.341 |
0.119 |
| 106 |
254645_at |
AT4G18520
|
SEL1, SEEDLING LETHAL 1 |
-0.337 |
-0.057 |
| 107 |
260636_at |
AT1G62430
|
ATCDS1, CDP-diacylglycerol synthase 1, CDS1, CDP-diacylglycerol synthase 1 |
-0.325 |
0.159 |
| 108 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
-0.315 |
0.033 |
| 109 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
-0.306 |
-0.130 |
| 110 |
258113_at |
AT3G14650
|
CYP72A11, cytochrome P450, family 72, subfamily A, polypeptide 11 |
-0.306 |
-0.138 |
| 111 |
249869_at |
AT5G23050
|
AAE17, acyl-activating enzyme 17 |
-0.305 |
-0.020 |
| 112 |
266225_at |
AT2G28900
|
ATOEP16-1, outer plastid envelope protein 16-1, ATOEP16-L, OUTER PLASTID ENVELOPE PROTEIN 16-L, OEP16, outer envelope protein 16, OEP16-1, outer plastid envelope protein 16-1 |
-0.300 |
-0.078 |
| 113 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.299 |
0.029 |
| 114 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
-0.297 |
0.017 |
| 115 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.290 |
-0.118 |
| 116 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
-0.284 |
0.019 |
| 117 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.282 |
-0.029 |
| 118 |
265078_at |
AT1G55500
|
ECT4, evolutionarily conserved C-terminal region 4 |
-0.280 |
0.018 |
| 119 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.273 |
-0.027 |
| 120 |
266439_s_at |
AT2G43200
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.272 |
-0.040 |
| 121 |
260364_at |
AT1G70560
|
CKRC1, cytokinin induced root curling 1, SAV3, SHADE AVOIDANCE 3, TAA1, tryptophan aminotransferase of Arabidopsis 1, WEI8, WEAK ETHYLENE INSENSITIVE 8 |
-0.270 |
-0.009 |
| 122 |
248502_at |
AT5G50450
|
[HCP-like superfamily protein with MYND-type zinc finger] |
-0.266 |
0.021 |
| 123 |
253351_at |
AT4G33700
|
unknown |
-0.259 |
0.124 |
| 124 |
262784_at |
AT1G10760
|
GWD, GWD1, SEX1, STARCH EXCESS 1, SOP, SOP1 |
-0.253 |
-0.103 |
| 125 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
-0.250 |
-0.137 |
| 126 |
254803_at |
AT4G13100
|
[RING/U-box superfamily protein] |
-0.250 |
-0.023 |
| 127 |
259014_at |
AT3G07320
|
[O-Glycosyl hydrolases family 17 protein] |
-0.249 |
0.118 |
| 128 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
-0.248 |
-0.067 |
| 129 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
-0.248 |
-0.048 |
| 130 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
-0.248 |
0.118 |
| 131 |
259600_at |
AT1G35220
|
unknown |
-0.247 |
0.052 |
| 132 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
-0.244 |
-0.076 |
| 133 |
253421_at |
AT4G32340
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.243 |
-0.124 |
| 134 |
246483_at |
AT5G16000
|
AtNIK1, NIK1, NSP-interacting kinase 1 |
-0.241 |
0.035 |
| 135 |
247744_at |
AT5G59020
|
unknown |
-0.240 |
0.073 |
| 136 |
257487_at |
AT1G71850
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
-0.239 |
0.137 |
| 137 |
246516_at |
AT5G15740
|
[O-fucosyltransferase family protein] |
-0.238 |
0.107 |
| 138 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
-0.238 |
0.088 |
| 139 |
259303_at |
AT3G05130
|
unknown |
-0.237 |
0.085 |
| 140 |
255931_at |
AT1G12710
|
AtPP2-A12, phloem protein 2-A12, PP2-A12, phloem protein 2-A12 |
-0.237 |
0.064 |
| 141 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
-0.235 |
0.082 |
| 142 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.233 |
-0.053 |
| 143 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.231 |
-0.062 |
| 144 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
-0.230 |
0.018 |
| 145 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.230 |
0.030 |
| 146 |
256822_at |
AT3G22190
|
IQD5, IQ-domain 5 |
-0.229 |
-0.019 |
| 147 |
249422_at |
AT5G39760
|
AtHB23, homeobox protein 23, HB23, homeobox protein 23, ZHD10, ZINC FINGER HOMEODOMAIN 10 |
-0.225 |
-0.023 |
| 148 |
254430_at |
AT4G20820
|
[FAD-binding Berberine family protein] |
-0.222 |
-0.033 |
| 149 |
266598_at |
AT2G46110
|
KPHMT1, ketopantoate hydroxymethyltransferase 1, PANB1 |
-0.220 |
0.004 |
| 150 |
263912_at |
AT2G36390
|
BE3, BRANCHING ENZYME 3, SBE2.1, starch branching enzyme 2.1 |
-0.217 |
-0.022 |
| 151 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
-0.216 |
-0.012 |