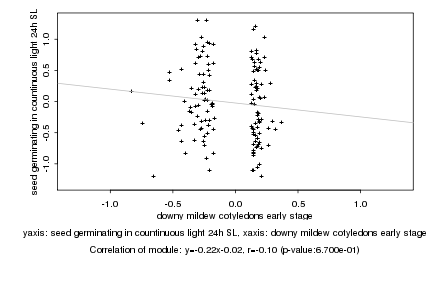
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
downy mildew cotyledons early stage |
Log2 signal ratio
seed germinating in countinuous light 24h SL |
| 1 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.361 |
-0.322 |
| 2 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
0.313 |
-0.435 |
| 3 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.296 |
-0.318 |
| 4 |
246968_at |
AT5G24870
|
[RING/U-box superfamily protein] |
0.274 |
0.292 |
| 5 |
246620_at |
AT5G36220
|
CYP81D1, cytochrome P450, family 81, subfamily D, polypeptide 1, CYP91A1, CYTOCHROME P450 91A1 |
0.260 |
-0.704 |
| 6 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
0.257 |
-0.432 |
| 7 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.236 |
0.510 |
| 8 |
252993_at |
AT4G38540
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.232 |
1.036 |
| 9 |
258037_at |
AT3G21230
|
4CL5, 4-coumarate:CoA ligase 5 |
0.230 |
0.079 |
| 10 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.230 |
0.714 |
| 11 |
252896_at |
AT4G39480
|
CYP96A9, cytochrome P450, family 96, subfamily A, polypeptide 9 |
0.207 |
-0.284 |
| 12 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
0.205 |
-1.192 |
| 13 |
259520_at |
AT1G12320
|
unknown |
0.205 |
-0.747 |
| 14 |
265931_at |
AT2G18520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.201 |
0.287 |
| 15 |
252677_at |
AT3G44320
|
AtNIT3, NITRILASE 3, NIT3, nitrilase 3 |
0.199 |
0.062 |
| 16 |
246282_at |
AT4G36580
|
[AAA-type ATPase family protein] |
0.197 |
0.636 |
| 17 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
0.192 |
-0.501 |
| 18 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.191 |
0.558 |
| 19 |
251895_at |
AT3G54420
|
ATCHITIV, CHITINASE CLASS IV, ATEP3, homolog of carrot EP3-3 chitinase, CHIV, EP3, homolog of carrot EP3-3 chitinase |
0.188 |
-0.319 |
| 20 |
261785_at |
AT1G08230
|
[Transmembrane amino acid transporter family protein] |
0.188 |
0.077 |
| 21 |
259399_at |
AT1G17710
|
AtPEPC1, Arabidopsis thaliana phosphoethanolamine/phosphocholine phosphatase 1, PEPC1, phosphoethanolamine/phosphocholine phosphatase 1 |
0.187 |
-0.326 |
| 22 |
266782_at |
AT2G29120
|
ATGLR2.7, glutamate receptor 2.7, GLR2.7, GLUTAMATE RECEPTOR 2.7, GLR2.7, glutamate receptor 2.7 |
0.186 |
-1.004 |
| 23 |
251896_at |
AT3G54390
|
[sequence-specific DNA binding transcription factors] |
0.185 |
-0.656 |
| 24 |
261023_at |
AT1G12200
|
FMO, flavin monooxygenase |
0.183 |
-0.689 |
| 25 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
0.181 |
0.509 |
| 26 |
261585_at |
AT1G01010
|
ANAC001, NAC domain containing protein 1, NAC001, NAC domain containing protein 1 |
0.179 |
-0.186 |
| 27 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
0.178 |
-0.286 |
| 28 |
255957_at |
AT1G22160
|
unknown |
0.177 |
0.683 |
| 29 |
262464_at |
AT1G50280
|
[Phototropic-responsive NPH3 family protein] |
0.174 |
-0.164 |
| 30 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
0.173 |
-0.212 |
| 31 |
254430_at |
AT4G20820
|
[FAD-binding Berberine family protein] |
0.173 |
-0.589 |
| 32 |
245855_at |
AT5G13550
|
SULTR4;1, sulfate transporter 4.1 |
0.173 |
-0.415 |
| 33 |
251472_at |
AT3G59580
|
NLP9, NIN-like protein 9 |
0.173 |
-1.045 |
| 34 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.172 |
-0.591 |
| 35 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
0.170 |
0.504 |
| 36 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.170 |
-0.720 |
| 37 |
245885_at |
AT5G09440
|
EXL4, EXORDIUM like 4 |
0.169 |
0.210 |
| 38 |
246133_at |
AT5G20960
|
AAO1, aldehyde oxidase 1, AO1, aldehyde oxidase 1, AOalpha, aldehyde oxidase alpha, AT-AO1, ARABIDOPSIS THALIANA ALDEHYDE OXIDASE 1, ATAO, AtAO1, Arabidopsis thaliana aldehyde oxidase 1 |
0.168 |
0.254 |
| 39 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
0.168 |
0.300 |
| 40 |
261168_at |
AT1G04945
|
[HIT-type Zinc finger family protein] |
0.166 |
0.191 |
| 41 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
0.165 |
0.777 |
| 42 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
0.163 |
0.519 |
| 43 |
259327_at |
AT3G16460
|
JAL34, jacalin-related lectin 34 |
0.162 |
0.820 |
| 44 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
0.162 |
-0.728 |
| 45 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.158 |
-0.341 |
| 46 |
252134_at |
AT3G50910
|
unknown |
0.158 |
1.215 |
| 47 |
251292_at |
AT3G61920
|
unknown |
0.157 |
-0.636 |
| 48 |
259503_at |
AT1G15870
|
[Mitochondrial glycoprotein family protein] |
0.156 |
0.240 |
| 49 |
253048_at |
AT4G37560
|
[Acetamidase/Formamidase family protein] |
0.154 |
-0.534 |
| 50 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
0.149 |
0.566 |
| 51 |
258009_at |
AT3G19440
|
[Pseudouridine synthase family protein] |
0.148 |
-0.045 |
| 52 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
0.146 |
0.627 |
| 53 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
0.146 |
0.339 |
| 54 |
263918_at |
AT2G36590
|
ATPROT3, PROLINE TRANSPORTER 3, ProT3, proline transporter 3 |
0.144 |
0.491 |
| 55 |
259746_at |
AT1G71060
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.144 |
-0.440 |
| 56 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
0.144 |
0.033 |
| 57 |
248961_at |
AT5G45650
|
[subtilase family protein] |
0.143 |
1.163 |
| 58 |
265433_at |
AT2G20950
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.143 |
-0.517 |
| 59 |
252736_at |
AT3G43210
|
ATNACK2, ARABIDOPSIS NPK1-ACTIVATING KINESIN 2, NACK2, NPK1-ACTIVATING KINESIN 2, TES, TETRASPORE |
0.142 |
-0.803 |
| 60 |
246120_at |
AT5G20360
|
Phox3, Phox3 |
0.141 |
-0.489 |
| 61 |
261106_at |
AT1G62990
|
IXR11, KNAT7, KNOTTED-like homeobox of Arabidopsis thaliana 7 |
0.141 |
-0.826 |
| 62 |
256981_at |
AT3G13380
|
BRL3, BRI1-like 3 |
0.139 |
-0.866 |
| 63 |
248939_at |
AT5G45790
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
0.139 |
-0.677 |
| 64 |
262480_at |
AT1G11340
|
[S-locus lectin protein kinase family protein] |
0.138 |
-0.786 |
| 65 |
253940_at |
AT4G26950
|
unknown |
0.137 |
-1.103 |
| 66 |
255622_at |
AT4G01070
|
GT72B1, UGT72B1, UDP-GLUCOSE-DEPENDENT GLUCOSYLTRANSFERASE 72 B1 |
0.133 |
0.683 |
| 67 |
256529_at |
AT1G33260
|
[Protein kinase superfamily protein] |
0.130 |
-1.104 |
| 68 |
264806_at |
AT1G08610
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.129 |
-0.426 |
| 69 |
264444_at |
AT1G27360
|
SPL11, squamosa promoter-like 11 |
0.125 |
0.282 |
| 70 |
246516_at |
AT5G15740
|
[O-fucosyltransferase family protein] |
0.125 |
0.814 |
| 71 |
266623_at |
AT2G35390
|
[Phosphoribosyltransferase family protein] |
0.123 |
0.125 |
| 72 |
248065_at |
AT5G55580
|
[Mitochondrial transcription termination factor family protein] |
0.122 |
0.718 |
| 73 |
254761_at |
AT4G13195
|
CLE44, CLAVATA3/ESR-RELATED 44 |
0.122 |
-0.398 |
| 74 |
267096_at |
AT2G38180
|
[SGNH hydrolase-type esterase superfamily protein] |
0.122 |
-0.027 |
| 75 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
-0.836 |
0.168 |
| 76 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
-0.743 |
-0.346 |
| 77 |
267459_at |
AT2G33850
|
unknown |
-0.659 |
-1.190 |
| 78 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.532 |
0.349 |
| 79 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-0.529 |
0.466 |
| 80 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.459 |
-0.464 |
| 81 |
258751_at |
AT3G05890
|
RCI2B, RARE-COLD-INDUCIBLE 2B |
-0.438 |
0.521 |
| 82 |
248509_at |
AT5G50335
|
unknown |
-0.434 |
-0.376 |
| 83 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
-0.432 |
-0.635 |
| 84 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
-0.408 |
0.002 |
| 85 |
247713_at |
AT5G59330
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.401 |
-0.820 |
| 86 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
-0.374 |
-0.145 |
| 87 |
254074_at |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
-0.360 |
-0.082 |
| 88 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
-0.357 |
0.218 |
| 89 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
-0.356 |
-0.162 |
| 90 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
-0.330 |
-0.355 |
| 91 |
245891_at |
AT5G09220
|
AAP2, amino acid permease 2 |
-0.329 |
-0.611 |
| 92 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
-0.325 |
-0.077 |
| 93 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.324 |
0.617 |
| 94 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.324 |
0.121 |
| 95 |
246215_at |
AT4G37180
|
[Homeodomain-like superfamily protein] |
-0.320 |
0.922 |
| 96 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-0.318 |
0.849 |
| 97 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
-0.307 |
1.316 |
| 98 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
-0.304 |
-0.237 |
| 99 |
250277_at |
AT5G12940
|
[Leucine-rich repeat (LRR) family protein] |
-0.300 |
0.194 |
| 100 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.297 |
0.713 |
| 101 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
-0.296 |
-0.059 |
| 102 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.290 |
0.440 |
| 103 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
-0.284 |
-0.444 |
| 104 |
248163_at |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
-0.281 |
0.730 |
| 105 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
-0.277 |
-0.426 |
| 106 |
250098_at |
AT5G17350
|
unknown |
-0.277 |
-0.307 |
| 107 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
-0.272 |
1.041 |
| 108 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.270 |
0.811 |
| 109 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
-0.269 |
0.135 |
| 110 |
258939_at |
AT3G10020
|
unknown |
-0.268 |
0.237 |
| 111 |
263265_at |
AT2G38820
|
unknown |
-0.260 |
-0.640 |
| 112 |
266831_at |
AT2G22830
|
SQE2, squalene epoxidase 2 |
-0.259 |
0.308 |
| 113 |
267393_at |
AT2G44500
|
[O-fucosyltransferase family protein] |
-0.257 |
0.445 |
| 114 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
-0.257 |
0.891 |
| 115 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
-0.255 |
0.031 |
| 116 |
248710_at |
AT5G48480
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
-0.254 |
0.142 |
| 117 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.252 |
0.232 |
| 118 |
260742_at |
AT1G15050
|
IAA34, indole-3-acetic acid inducible 34 |
-0.249 |
-0.561 |
| 119 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
-0.248 |
-0.701 |
| 120 |
261203_at |
AT1G12845
|
unknown |
-0.244 |
0.042 |
| 121 |
247126_at |
AT5G66080
|
APD9, Arabidopsis Pp2c clade D 9 |
-0.241 |
-0.291 |
| 122 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-0.239 |
-0.914 |
| 123 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
-0.238 |
1.312 |
| 124 |
263799_at |
AT2G24550
|
unknown |
-0.231 |
0.949 |
| 125 |
259979_at |
AT1G76600
|
unknown |
-0.229 |
-0.501 |
| 126 |
255285_at |
AT4G04630
|
unknown |
-0.226 |
0.183 |
| 127 |
258376_at |
AT3G17680
|
[Kinase interacting (KIP1-like) family protein] |
-0.225 |
0.738 |
| 128 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.224 |
0.027 |
| 129 |
247878_at |
AT5G57760
|
unknown |
-0.224 |
-0.150 |
| 130 |
252050_at |
AT3G52550
|
unknown |
-0.223 |
-0.377 |
| 131 |
261745_at |
AT1G08500
|
AtENODL18, ENODL18, early nodulin-like protein 18 |
-0.222 |
0.597 |
| 132 |
259015_at |
AT3G07350
|
unknown |
-0.219 |
-0.384 |
| 133 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-0.219 |
0.513 |
| 134 |
258275_at |
AT3G15760
|
unknown |
-0.214 |
-0.293 |
| 135 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
-0.214 |
0.190 |
| 136 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
-0.213 |
-1.096 |
| 137 |
258641_at |
AT3G08030
|
unknown |
-0.208 |
0.936 |
| 138 |
257076_at |
AT3G19680
|
unknown |
-0.208 |
0.421 |
| 139 |
257679_at |
AT3G20470
|
ATGRP-5, ATGRP5, GRP-5, GLYCINE-RICH PROTEIN 5, GRP5, glycine-rich protein 5 |
-0.198 |
-0.035 |
| 140 |
267529_at |
AT2G45490
|
AtAUR3, ataurora3, AUR3, ataurora3 |
-0.191 |
-0.042 |
| 141 |
247597_at |
AT5G60860
|
AtRABA1f, RAB GTPase homolog A1F, RABA1f, RAB GTPase homolog A1F |
-0.189 |
-0.073 |
| 142 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.186 |
-0.024 |
| 143 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
-0.181 |
-0.828 |
| 144 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
-0.178 |
-0.446 |
| 145 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
-0.177 |
0.620 |
| 146 |
260944_at |
AT1G45130
|
AtBGAL5, BGAL5, beta-galactosidase 5 |
-0.177 |
0.926 |
| 147 |
249117_at |
AT5G43840
|
AT-HSFA6A, heat shock transcription factor A6A, HSFA6A, heat shock transcription factor A6A |
-0.175 |
-0.261 |