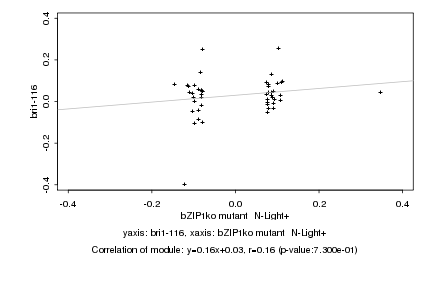
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
bZIP1ko mutant N-Light+ |
Log2 signal ratio
bri1-116 |
| 1 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
0.347 |
0.046 |
| 2 |
266843_at |
AT2G26135
|
[RING/U-box protein with C6HC-type zinc finger] |
0.112 |
0.097 |
| 3 |
264997_at |
AT1G67220
|
ATHPCAT1, ARABIDOPSIS THALIANA P300/CBP ACETYLTRANSFERASE-RELATED PROTEIN 1, HAC02, HAC2, histone acetyltransferase of the CBP family 2 |
0.109 |
0.095 |
| 4 |
253718_at |
AT4G29450
|
[Leucine-rich repeat protein kinase family protein] |
0.107 |
0.030 |
| 5 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
0.107 |
0.007 |
| 6 |
252237_at |
AT3G49950
|
[GRAS family transcription factor] |
0.103 |
0.255 |
| 7 |
246896_at |
AT5G25550
|
[Leucine-rich repeat (LRR) family protein] |
0.100 |
0.089 |
| 8 |
260187_at |
AT1G36000
|
LBD5, LOB domain-containing protein 5 |
0.092 |
0.014 |
| 9 |
266200_at |
AT2G38920
|
[SPX (SYG1/Pho81/XPR1) domain-containing protein / zinc finger (C3HC4-type RING finger) protein-related] |
0.091 |
-0.009 |
| 10 |
261260_at |
AT1G26680
|
[transcriptional factor B3 family protein] |
0.091 |
0.049 |
| 11 |
248948_at |
AT5G45560
|
[Pleckstrin homology (PH) domain-containing protein / lipid-binding START domain-containing protein] |
0.089 |
-0.029 |
| 12 |
261499_at |
AT1G28430
|
CYP705A24, cytochrome P450, family 705, subfamily A, polypeptide 24 |
0.088 |
0.023 |
| 13 |
249639_at |
AT5G36930
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.084 |
0.131 |
| 14 |
256188_at |
AT1G30160
|
unknown |
0.084 |
0.044 |
| 15 |
258596_at |
AT3G04510
|
LSH2, LIGHT SENSITIVE HYPOCOTYLS 2 |
0.084 |
0.030 |
| 16 |
266602_at |
AT2G46050
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.079 |
0.085 |
| 17 |
249223_at |
AT5G42120
|
LecRK-S.6, L-type lectin receptor kinase S.6 |
0.079 |
-0.030 |
| 18 |
257689_at |
AT3G12820
|
AtMYB10, myb domain protein 10, MYB10, myb domain protein 10 |
0.077 |
0.074 |
| 19 |
263144_at |
AT1G54070
|
[Dormancy/auxin associated family protein] |
0.077 |
0.046 |
| 20 |
248887_at |
AT5G46115
|
unknown |
0.076 |
-0.010 |
| 21 |
251754_at |
AT3G55780
|
[Glycosyl hydrolase superfamily protein] |
0.076 |
0.012 |
| 22 |
252299_at |
AT3G49150
|
[F-box/RNI-like superfamily protein] |
0.076 |
-0.002 |
| 23 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.075 |
-0.050 |
| 24 |
261752_at |
AT1G76290
|
[AMP-dependent synthetase and ligase family protein] |
0.073 |
0.093 |
| 25 |
259399_at |
AT1G17710
|
AtPEPC1, Arabidopsis thaliana phosphoethanolamine/phosphocholine phosphatase 1, PEPC1, phosphoethanolamine/phosphocholine phosphatase 1 |
0.073 |
0.038 |
| 26 |
262075_at |
AT1G59560
|
DAL2, DIAP1-like protein 2, SPL1, SP1-like 1, ZCF61 |
-0.148 |
0.083 |
| 27 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
-0.124 |
-0.394 |
| 28 |
266492_at |
AT2G07020
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.115 |
0.081 |
| 29 |
260201_at |
AT1G67600
|
[Acid phosphatase/vanadium-dependent haloperoxidase-related protein] |
-0.114 |
0.073 |
| 30 |
254437_s_at |
AT3G06433
|
[pseudogene] |
-0.111 |
0.045 |
| 31 |
246136_at |
AT5G28470
|
[Major facilitator superfamily protein] |
-0.105 |
0.042 |
| 32 |
254115_at |
AT4G24710
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.103 |
-0.045 |
| 33 |
264243_at |
AT1G54650
|
[Methyltransferase family protein] |
-0.102 |
0.020 |
| 34 |
266034_at |
AT2G06005
|
FIP1, FRIGIDA interacting protein 1 |
-0.100 |
0.002 |
| 35 |
265916_at |
AT3G31920
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 3.0e-163 P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
-0.100 |
0.080 |
| 36 |
260797_at |
AT1G78390
|
ATNCED9, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 9, NCED9, nine-cis-epoxycarotenoid dioxygenase 9 |
-0.098 |
-0.102 |
| 37 |
248897_at |
AT5G46360
|
ATKCO3, CA2+ ACTIVATED OUTWARD RECTIFYING K+ CHANNEL 3, KCO3, Ca2+ activated outward rectifying K+ channel 3 |
-0.090 |
-0.085 |
| 38 |
248928_at |
AT5G45970
|
ARAC2, Arabidopsis RAC-like 2, ATRAC2, ARABIDOPSIS THALIANA RAC 2, ATROP7, RHO-RELATED PROTEIN FROM PLANTS 7, RAC2, RAC-like 2, ROP7 |
-0.089 |
-0.042 |
| 39 |
255931_at |
AT1G12710
|
AtPP2-A12, phloem protein 2-A12, PP2-A12, phloem protein 2-A12 |
-0.089 |
0.058 |
| 40 |
259262_at |
AT3G01020
|
ATISU2, ISCU-LIKE 2, ISU2, ISCU-like 2 |
-0.085 |
0.143 |
| 41 |
247995_at |
AT5G56160
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.083 |
0.057 |
| 42 |
262688_at |
AT1G62680
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.083 |
0.021 |
| 43 |
245297_at |
AT4G16510
|
[YbaK/aminoacyl-tRNA synthetase-associated domain] |
-0.083 |
0.055 |
| 44 |
249430_at |
AT5G39900
|
[Small GTP-binding protein] |
-0.082 |
-0.016 |
| 45 |
267217_at |
AT2G02610
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.082 |
0.035 |
| 46 |
255232_at |
AT4G05330
|
AGD13, ARF-GAP domain 13 |
-0.081 |
0.254 |
| 47 |
246561_at |
AT5G15570
|
[Bromodomain transcription factor] |
-0.080 |
0.051 |
| 48 |
245490_at |
AT4G16310
|
LDL3, LSD1-like 3 |
-0.080 |
-0.100 |
| 49 |
258068_at |
AT3G25990
|
[Homeodomain-like superfamily protein] |
-0.079 |
0.049 |