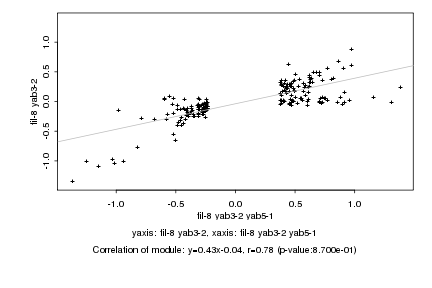
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
fil-8 yab3-2 yab5-1 |
Log2 signal ratio
fil-8 yab3-2 |
| 1 |
256021_at |
AT1G58270
|
ZW9 |
1.381 |
0.244 |
| 2 |
265611_at |
AT2G25510
|
unknown |
1.305 |
-0.008 |
| 3 |
265615_at |
AT2G25450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.156 |
0.077 |
| 4 |
262010_at |
AT1G35612
|
[expressed protein] |
0.972 |
0.891 |
| 5 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.966 |
0.619 |
| 6 |
248944_at |
AT5G45500
|
[RNI-like superfamily protein] |
0.956 |
0.031 |
| 7 |
262206_at |
AT2G01090
|
[Ubiquinol-cytochrome C reductase hinge protein] |
0.913 |
-0.008 |
| 8 |
252703_at |
AT3G43740
|
[Leucine-rich repeat (LRR) family protein] |
0.909 |
0.155 |
| 9 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.904 |
0.559 |
| 10 |
265051_at |
AT1G52100
|
[Mannose-binding lectin superfamily protein] |
0.892 |
-0.036 |
| 11 |
252659_at |
AT3G44430
|
unknown |
0.877 |
0.073 |
| 12 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
0.859 |
0.683 |
| 13 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
0.855 |
-0.010 |
| 14 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.820 |
0.390 |
| 15 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
0.800 |
0.371 |
| 16 |
260312_at |
AT1G63880
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.767 |
0.033 |
| 17 |
246025_at |
AT5G21150
|
AGO9, ARGONAUTE 9 |
0.765 |
0.570 |
| 18 |
248016_at |
AT5G56380
|
[F-box/RNI-like/FBD-like domains-containing protein] |
0.748 |
0.060 |
| 19 |
251347_at |
AT3G61010
|
[Ferritin/ribonucleotide reductase-like family protein] |
0.742 |
0.065 |
| 20 |
261260_at |
AT1G26680
|
[transcriptional factor B3 family protein] |
0.730 |
0.363 |
| 21 |
249791_at |
AT5G23810
|
AAP7, amino acid permease 7 |
0.728 |
0.078 |
| 22 |
252648_at |
AT3G44630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.723 |
-0.010 |
| 23 |
248079_at |
AT5G55790
|
unknown |
0.721 |
-0.022 |
| 24 |
247988_at |
AT5G56910
|
[Proteinase inhibitor I25, cystatin, conserved region] |
0.707 |
0.064 |
| 25 |
248847_at |
AT5G46510
|
VICTL, VARIATION IN COMPOUND TRIGGERED ROOT growth response-like |
0.704 |
0.011 |
| 26 |
264497_at |
AT1G30840
|
ATPUP4, purine permease 4, PUP4, purine permease 4 |
0.704 |
0.448 |
| 27 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
0.704 |
0.497 |
| 28 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
0.698 |
-0.006 |
| 29 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
0.675 |
0.490 |
| 30 |
256621_at |
AT3G24450
|
[Heavy metal transport/detoxification superfamily protein ] |
0.651 |
0.497 |
| 31 |
247196_at |
AT5G65510
|
AIL7, AINTEGUMENTA-like 7, PLT7, PLETHORA 7 |
0.645 |
0.329 |
| 32 |
257460_at |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
0.632 |
0.395 |
| 33 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
0.622 |
0.382 |
| 34 |
263380_at |
AT2G40200
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.622 |
0.329 |
| 35 |
257035_at |
AT3G19270
|
CYP707A4, cytochrome P450, family 707, subfamily A, polypeptide 4 |
0.620 |
0.269 |
| 36 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
0.619 |
0.451 |
| 37 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.617 |
0.414 |
| 38 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.615 |
0.325 |
| 39 |
250931_at |
AT5G03200
|
LUL1, LOG2-LIKE UBIQUITIN LIGASE1 |
0.612 |
0.042 |
| 40 |
256987_at |
AT3G28560
|
[BCS1 AAA-type ATPase] |
0.612 |
0.157 |
| 41 |
262908_at |
AT1G59900
|
AT-E1 ALPHA, pyruvate dehydrogenase complex E1 alpha subunit, E1 ALPHA, pyruvate dehydrogenase complex E1 alpha subunit |
0.611 |
0.249 |
| 42 |
252485_at |
AT3G46530
|
RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13 |
0.602 |
-0.055 |
| 43 |
246485_at |
AT5G16080
|
AtCXE17, carboxyesterase 17, CXE17, carboxyesterase 17 |
0.599 |
0.004 |
| 44 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.584 |
0.279 |
| 45 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
0.584 |
0.117 |
| 46 |
251254_at |
AT3G62270
|
BOR2, REQUIRES HIGH BORON 2 |
0.577 |
0.239 |
| 47 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
0.569 |
0.180 |
| 48 |
250868_at |
AT5G03860
|
MLS, malate synthase |
0.565 |
0.316 |
| 49 |
246079_at |
AT5G20450
|
unknown |
0.555 |
0.031 |
| 50 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.553 |
0.061 |
| 51 |
250063_at |
AT5G17880
|
CSA1, constitutive shade-avoidance1 |
0.540 |
0.059 |
| 52 |
264741_at |
AT1G62290
|
[Saposin-like aspartyl protease family protein] |
0.533 |
0.382 |
| 53 |
251062_at |
AT5G01840
|
ATOFP1, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 1, OFP1, ovate family protein 1 |
0.528 |
0.254 |
| 54 |
264156_at |
AT1G65280
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.519 |
-0.019 |
| 55 |
264824_at |
AT1G03420
|
Sadhu4-2, sadhu non-coding retrotransposon 4-2 |
0.498 |
0.467 |
| 56 |
262913_at |
AT1G59960
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.497 |
0.084 |
| 57 |
246478_at |
AT5G15980
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.494 |
0.191 |
| 58 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.493 |
0.360 |
| 59 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.493 |
0.171 |
| 60 |
255900_at |
AT1G17830
|
unknown |
0.485 |
0.348 |
| 61 |
250755_at |
AT5G05750
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.482 |
0.006 |
| 62 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.480 |
0.236 |
| 63 |
256457_at |
AT1G75230
|
[DNA glycosylase superfamily protein] |
0.479 |
0.013 |
| 64 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.473 |
0.021 |
| 65 |
261496_at |
AT1G28360
|
ATERF12, ERF DOMAIN PROTEIN 12, ERF12, ERF domain protein 12 |
0.470 |
0.305 |
| 66 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.469 |
0.253 |
| 67 |
250751_at |
AT5G05890
|
[UDP-Glycosyltransferase superfamily protein] |
0.466 |
0.106 |
| 68 |
249340_at |
AT5G41140
|
[Myosin heavy chain-related protein] |
0.463 |
-0.051 |
| 69 |
252700_at |
AT3G43690
|
[copia-like retrotransposon family protein, has a 1.4e-29 P-value blast match to gb|AAG52950.1| putative envelope protein (Endovir1-1) (Arabidopsis thaliana) (Ty1_Copia-family)] |
0.463 |
-0.013 |
| 70 |
262039_at |
AT1G80050
|
APT2, adenine phosphoribosyl transferase 2, ATAPT2, PHT1.1 |
0.456 |
0.260 |
| 71 |
249124_at |
AT5G43740
|
[Disease resistance protein (CC-NBS-LRR class) family] |
0.456 |
0.050 |
| 72 |
257026_at |
AT3G19200
|
unknown |
0.455 |
0.295 |
| 73 |
267627_at |
AT2G42270
|
[U5 small nuclear ribonucleoprotein helicase] |
0.451 |
-0.022 |
| 74 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.451 |
-0.037 |
| 75 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.451 |
0.185 |
| 76 |
256927_at |
AT3G22550
|
unknown |
0.446 |
0.298 |
| 77 |
255414_at |
AT4G03156
|
[small GTPase-related] |
0.438 |
0.627 |
| 78 |
246133_at |
AT5G20960
|
AAO1, aldehyde oxidase 1, AO1, aldehyde oxidase 1, AOalpha, aldehyde oxidase alpha, AT-AO1, ARABIDOPSIS THALIANA ALDEHYDE OXIDASE 1, ATAO, AtAO1, Arabidopsis thaliana aldehyde oxidase 1 |
0.433 |
0.230 |
| 79 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.432 |
0.294 |
| 80 |
250509_at |
AT5G09970
|
CYP78A7, cytochrome P450, family 78, subfamily A, polypeptide 7 |
0.426 |
0.220 |
| 81 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.425 |
0.149 |
| 82 |
261224_at |
AT1G20160
|
ATSBT5.2 |
0.421 |
0.264 |
| 83 |
260632_at |
AT1G62360
|
BUM, BUMBERSHOOT, BUM1, BUMBERSHOOT 1, SHL, SHOOTLESS, STM, SHOOT MERISTEMLESS, WAM, WALDMEISTER, WAM1, WALDMEISTER 1 |
0.420 |
0.247 |
| 84 |
253084_at |
AT4G36260
|
SRS2, SHI RELATED SEQUENCE 2, STY2, STYLISH 2 |
0.417 |
0.368 |
| 85 |
262041_at |
AT1G80100
|
AHP6, histidine phosphotransfer protein 6, HP6, histidine phosphotransfer protein 6 |
0.415 |
0.182 |
| 86 |
266814_at |
AT2G44910
|
ATHB-4, ARABIDOPSIS THALIANA HOMEOBOX-LEUCINE ZIPPER PROTEIN 4, ATHB4, homeobox-leucine zipper protein 4, HB4, homeobox-leucine zipper protein 4 |
0.411 |
0.196 |
| 87 |
246393_at |
AT1G58150
|
unknown |
0.405 |
0.010 |
| 88 |
265276_at |
AT2G28400
|
unknown |
0.405 |
0.312 |
| 89 |
260484_at |
AT1G68360
|
[C2H2 and C2HC zinc fingers superfamily protein] |
0.402 |
0.264 |
| 90 |
267428_at |
AT2G34840
|
[Coatomer epsilon subunit] |
0.401 |
0.013 |
| 91 |
248954_at |
AT5G45420
|
maMYB, membrane anchored MYB |
0.400 |
0.034 |
| 92 |
246888_at |
AT5G26270
|
unknown |
0.396 |
0.017 |
| 93 |
249112_at |
AT5G43780
|
APS4 |
0.393 |
0.180 |
| 94 |
251983_at |
AT3G53210
|
UMAMIT6, Usually multiple acids move in and out Transporters 6 |
0.385 |
0.297 |
| 95 |
252654_at |
AT3G44740
|
[Class II aaRS and biotin synthetases superfamily protein] |
0.384 |
0.030 |
| 96 |
250446_at |
AT5G10770
|
[Eukaryotic aspartyl protease family protein] |
0.384 |
0.108 |
| 97 |
245341_at |
AT4G16447
|
unknown |
0.382 |
0.273 |
| 98 |
248722_at |
AT5G47810
|
PFK2, phosphofructokinase 2 |
0.381 |
-0.024 |
| 99 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
0.379 |
0.370 |
| 100 |
267096_at |
AT2G38180
|
[SGNH hydrolase-type esterase superfamily protein] |
0.377 |
0.150 |
| 101 |
262446_at |
AT1G49310
|
unknown |
0.377 |
0.323 |
| 102 |
259796_at |
AT1G64270
|
[Mutator-like transposase family, has a 1.0e-06 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
0.374 |
-0.038 |
| 103 |
249331_at |
AT5G40950
|
RPL27, ribosomal protein large subunit 27 |
0.374 |
0.021 |
| 104 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-1.369 |
-1.341 |
| 105 |
254327_at |
AT4G22490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.253 |
-1.001 |
| 106 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
-1.151 |
-1.090 |
| 107 |
247713_at |
AT5G59330
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.032 |
-0.975 |
| 108 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
-1.016 |
-1.030 |
| 109 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.988 |
-0.151 |
| 110 |
254452_at |
AT4G21100
|
DDB1B, damaged DNA binding protein 1B |
-0.945 |
-1.001 |
| 111 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
-0.823 |
-0.760 |
| 112 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.790 |
-0.281 |
| 113 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.687 |
-0.291 |
| 114 |
267376_at |
AT2G26330
|
ER, ERECTA, QRP1, QUANTITATIVE RESISTANCE TO PLECTOSPHAERELLA 1 |
-0.600 |
0.047 |
| 115 |
250135_at |
AT5G15360
|
unknown |
-0.595 |
0.067 |
| 116 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.579 |
-0.293 |
| 117 |
259801_at |
AT1G72230
|
[Cupredoxin superfamily protein] |
-0.570 |
-0.205 |
| 118 |
252663_at |
AT3G44070
|
[Glycosyl hydrolase family 35 protein] |
-0.559 |
0.092 |
| 119 |
248695_at |
AT5G48350
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
-0.530 |
-0.050 |
| 120 |
262421_at |
AT1G50290
|
unknown |
-0.527 |
0.066 |
| 121 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.522 |
-0.194 |
| 122 |
245138_at |
AT2G45190
|
AFO, ABNORMAL FLORAL ORGANS, FIL, FILAMENTOUS FLOWER, YAB1, YABBY1 |
-0.520 |
-0.545 |
| 123 |
255709_at |
AT4G00180
|
YAB3, YABBY3 |
-0.507 |
-0.640 |
| 124 |
253514_at |
AT4G31805
|
POLAR, POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION |
-0.494 |
-0.131 |
| 125 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.487 |
-0.052 |
| 126 |
250828_at |
AT5G05250
|
unknown |
-0.486 |
-0.400 |
| 127 |
266824_at |
AT2G22800
|
HAT9 |
-0.484 |
-0.338 |
| 128 |
265194_at |
AT1G05010
|
ACO4, ethylene forming enzyme, EAT1, EFE, ethylene-forming enzyme |
-0.464 |
-0.305 |
| 129 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
-0.464 |
-0.123 |
| 130 |
250566_at |
AT5G08070
|
TCP17, TCP domain protein 17 |
-0.460 |
-0.257 |
| 131 |
247592_at |
AT5G60780
|
ATNRT2.3, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 2.3, NRT2.3, nitrate transporter 2.3 |
-0.459 |
-0.390 |
| 132 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.444 |
-0.355 |
| 133 |
260396_at |
AT1G69720
|
HO3, heme oxygenase 3 |
-0.444 |
-0.112 |
| 134 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.431 |
0.037 |
| 135 |
260776_at |
AT1G14580
|
[C2H2-like zinc finger protein] |
-0.429 |
-0.131 |
| 136 |
267317_at |
AT2G34700
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.427 |
-0.298 |
| 137 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.413 |
-0.141 |
| 138 |
260034_at |
AT1G68810
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.412 |
-0.220 |
| 139 |
252916_at |
AT4G38950
|
[ATP binding microtubule motor family protein] |
-0.410 |
-0.211 |
| 140 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
-0.405 |
-0.179 |
| 141 |
245884_at |
AT5G09300
|
[Thiamin diphosphate-binding fold (THDP-binding) superfamily protein] |
-0.405 |
-0.117 |
| 142 |
252280_at |
AT3G49260
|
iqd21, IQ-domain 21 |
-0.399 |
-0.216 |
| 143 |
262121_at |
AT1G02800
|
ATCEL2, cellulase 2, CEL2, CEL2, cellulase 2 |
-0.397 |
-0.192 |
| 144 |
257267_at |
AT3G15030
|
MEE35, maternal effect embryo arrest 35, TCP4, TCP family transcription factor 4 |
-0.395 |
-0.252 |
| 145 |
251704_at |
AT3G56360
|
unknown |
-0.377 |
-0.185 |
| 146 |
246346_at |
AT3G56810
|
unknown |
-0.374 |
-0.101 |
| 147 |
265960_at |
AT2G37470
|
[Histone superfamily protein] |
-0.372 |
-0.067 |
| 148 |
252691_at |
AT3G44050
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.371 |
-0.167 |
| 149 |
258530_at |
AT3G06840
|
unknown |
-0.365 |
-0.148 |
| 150 |
254670_at |
AT4G18390
|
TCP2, TCP2, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 2 |
-0.360 |
-0.203 |
| 151 |
255422_at |
AT4G03270
|
CYCD6;1, Cyclin D6;1 |
-0.352 |
-0.251 |
| 152 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
-0.347 |
-0.242 |
| 153 |
248148_at |
AT5G54930
|
[AT hook motif-containing protein] |
-0.326 |
-0.060 |
| 154 |
257714_at |
AT3G27360
|
H3.1, histone 3.1 |
-0.318 |
-0.130 |
| 155 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
-0.317 |
-0.209 |
| 156 |
247977_at |
AT5G56850
|
unknown |
-0.316 |
-0.188 |
| 157 |
266402_at |
AT2G38780
|
unknown |
-0.315 |
-0.090 |
| 158 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
-0.314 |
-0.244 |
| 159 |
248751_at |
AT5G47540
|
[Mo25 family protein] |
-0.313 |
-0.050 |
| 160 |
259686_at |
AT1G63100
|
[GRAS family transcription factor] |
-0.313 |
-0.120 |
| 161 |
254208_at |
AT4G24175
|
unknown |
-0.311 |
0.054 |
| 162 |
247599_at |
AT5G60880
|
BASL, BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE |
-0.308 |
-0.081 |
| 163 |
254639_at |
AT4G18750
|
DOT4, DEFECTIVELY ORGANIZED TRIBUTARIES 4 |
-0.308 |
0.038 |
| 164 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-0.303 |
-0.133 |
| 165 |
259945_at |
AT1G71460
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.289 |
-0.064 |
| 166 |
252628_at |
AT3G44960
|
unknown |
-0.286 |
-0.163 |
| 167 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.284 |
-0.152 |
| 168 |
260065_at |
AT1G73760
|
[RING/U-box superfamily protein] |
-0.283 |
-0.214 |
| 169 |
246557_at |
AT5G15510
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.282 |
-0.125 |
| 170 |
251178_at |
AT3G63440
|
ATCKX6, CYTOKININ OXIDASE 6, ATCKX7, CKX6, cytokinin oxidase/dehydrogenase 6 |
-0.281 |
-0.172 |
| 171 |
259611_at |
AT1G52280
|
AtRABG3d, RAB GTPase homolog G3D, RABG3d, RAB GTPase homolog G3D |
-0.278 |
-0.039 |
| 172 |
266867_at |
AT2G45770
|
CPFTSY, FRD4, FERRIC CHELATE REDUCTASE DEFECTIVE 4 |
-0.275 |
-0.039 |
| 173 |
246344_at |
AT3G56730
|
[Putative endonuclease or glycosyl hydrolase] |
-0.272 |
-0.215 |
| 174 |
257866_at |
AT3G17770
|
[Dihydroxyacetone kinase] |
-0.270 |
-0.089 |
| 175 |
264082_at |
AT2G28570
|
unknown |
-0.269 |
-0.170 |
| 176 |
250346_at |
AT5G11950
|
LOG8, LONELY GUY 8 |
-0.269 |
-0.109 |
| 177 |
264025_at |
AT2G21050
|
LAX2, like AUXIN RESISTANT 2 |
-0.266 |
-0.123 |
| 178 |
257903_at |
AT3G28460
|
[methyltransferases] |
-0.264 |
-0.090 |
| 179 |
267193_at |
AT2G30900
|
TBL43, TRICHOME BIREFRINGENCE-LIKE 43 |
-0.263 |
-0.147 |
| 180 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
-0.263 |
-0.114 |
| 181 |
264698_at |
AT1G70200
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.263 |
-0.029 |
| 182 |
258907_at |
AT3G06370
|
ATNHX4, NHX4, sodium hydrogen exchanger 4 |
-0.259 |
-0.147 |
| 183 |
261333_at |
AT1G44910
|
ATPRP40A, pre-mRNA-processing protein 40A, PRP40A, pre-mRNA-processing protein 40A |
-0.258 |
-0.089 |
| 184 |
256623_at |
AT3G19960
|
ATM1, myosin 1 |
-0.257 |
-0.167 |
| 185 |
255734_at |
AT1G25550
|
[myb-like transcription factor family protein] |
-0.256 |
-0.047 |
| 186 |
247118_at |
AT5G65890
|
ACR1, ACT domain repeat 1 |
-0.254 |
-0.138 |
| 187 |
250395_at |
AT5G10950
|
[Tudor/PWWP/MBT superfamily protein] |
-0.253 |
-0.265 |
| 188 |
261196_at |
AT1G12860
|
ICE2, INDUCER OF CBF EXPRESSION 2, SCRM2, SCREAM 2 |
-0.251 |
-0.101 |
| 189 |
254727_at |
AT4G13670
|
PTAC5, plastid transcriptionally active 5 |
-0.249 |
-0.029 |
| 190 |
248863_at |
AT5G46750
|
AGD9, ARF-GAP domain 9 |
-0.248 |
-0.145 |
| 191 |
250699_at |
AT5G06820
|
SRF2, STRUBBELIG-receptor family 2 |
-0.248 |
-0.016 |
| 192 |
251631_at |
AT3G57430
|
OTP84, ORGANELLE TRANSCRIPT PROCESSING 84 |
-0.246 |
-0.041 |
| 193 |
260457_at |
AT1G72480
|
[Lung seven transmembrane receptor family protein] |
-0.246 |
0.036 |
| 194 |
250519_at |
AT5G08460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.245 |
-0.071 |
| 195 |
245413_at |
AT4G17300
|
ATNS1, NS1, OVA8, ovule abortion 8 |
-0.245 |
-0.054 |
| 196 |
250087_at |
AT5G17270
|
[Protein prenylyltransferase superfamily protein] |
-0.244 |
-0.129 |
| 197 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.244 |
-0.083 |
| 198 |
251807_at |
AT3G55400
|
OVA1, OVULE ABORTION 1 |
-0.243 |
-0.073 |
| 199 |
258098_at |
AT3G23670
|
KINESIN-12B, PAKRP1L |
-0.242 |
-0.111 |
| 200 |
263322_at |
AT2G04270
|
RNE, RNase E, RNEE/G, RNAse E/G-like |
-0.242 |
-0.115 |
| 201 |
246069_at |
AT5G20220
|
[zinc knuckle (CCHC-type) family protein] |
-0.241 |
-0.115 |
| 202 |
259281_at |
AT3G01140
|
AtMYB106, myb domain protein 106, MYB106, myb domain protein 106, NOK, NOECK |
-0.240 |
-0.073 |
| 203 |
245876_at |
AT1G26230
|
Cpn60beta4, chaperonin-60beta4 |
-0.240 |
-0.001 |
| 204 |
264767_at |
AT1G61380
|
SD1-29, S-domain-1 29 |
-0.240 |
-0.071 |
| 205 |
253251_at |
AT4G34730
|
RBF1, RbfA domain-containing protein 1 |
-0.239 |
-0.076 |