|
probeID |
AGICode |
Annotation |
Log2 signal ratio
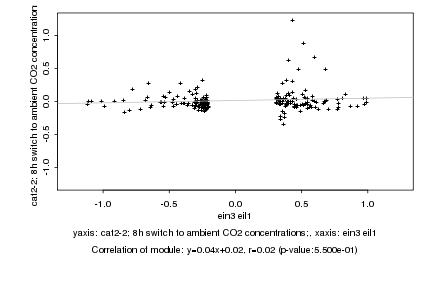
ein3 eil1 |
Log2 signal ratio
cat2-2; 8h switch to ambient CO2 concentrations; |
| 1 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
0.985 |
-0.005 |
| 2 |
256382_at |
AT1G66860
|
[Class I glutamine amidotransferase-like superfamily protein] |
0.983 |
0.046 |
| 3 |
256878_at |
AT3G26460
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.974 |
-0.033 |
| 4 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.960 |
0.053 |
| 5 |
249202_at |
AT5G42580
|
CYP705A12, cytochrome P450, family 705, subfamily A, polypeptide 12 |
0.917 |
-0.070 |
| 6 |
245689_at |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
0.864 |
-0.067 |
| 7 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.824 |
0.116 |
| 8 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.802 |
0.053 |
| 9 |
249205_at |
AT5G42600
|
MRN1, marneral synthase 1 |
0.777 |
-0.024 |
| 10 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.775 |
-0.084 |
| 11 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
0.771 |
0.043 |
| 12 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
0.765 |
-0.114 |
| 13 |
259866_at |
AT1G76640
|
CML39, CALMODULIN LIKE 39 |
0.699 |
-0.121 |
| 14 |
253070_at |
AT4G37850
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.687 |
0.024 |
| 15 |
255811_at |
AT4G10250
|
ATHSP22.0 |
0.677 |
0.004 |
| 16 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.673 |
0.490 |
| 17 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
0.670 |
0.004 |
| 18 |
264940_at |
AT1G60470
|
AtGolS4, galactinol synthase 4, GolS4, galactinol synthase 4 |
0.663 |
-0.023 |
| 19 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.622 |
-0.107 |
| 20 |
256527_at |
AT1G66100
|
[Plant thionin] |
0.605 |
-0.001 |
| 21 |
255527_at |
AT4G02360
|
unknown |
0.598 |
-0.088 |
| 22 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.596 |
0.015 |
| 23 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
0.592 |
0.682 |
| 24 |
256442_at |
AT3G10930
|
unknown |
0.582 |
0.091 |
| 25 |
247727_at |
AT5G59490
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.575 |
0.029 |
| 26 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
0.570 |
-0.084 |
| 27 |
266828_at |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
0.563 |
-0.046 |
| 28 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
0.556 |
-0.079 |
| 29 |
251480_at |
AT3G59710
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.543 |
-0.098 |
| 30 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.542 |
0.058 |
| 31 |
249203_at |
AT5G42590
|
CYP71A16, cytochrome P450, family 71, subfamily A, polypeptide 16, MRO, marneral oxidase |
0.540 |
-0.004 |
| 32 |
262659_at |
AT1G14240
|
[GDA1/CD39 nucleoside phosphatase family protein] |
0.538 |
-0.044 |
| 33 |
250464_at |
AT5G10040
|
unknown |
0.537 |
-0.000 |
| 34 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
0.529 |
0.046 |
| 35 |
264245_at |
AT1G60450
|
AtGolS7, galactinol synthase 7, GolS7, galactinol synthase 7 |
0.529 |
-0.038 |
| 36 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.528 |
0.181 |
| 37 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.525 |
-0.017 |
| 38 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
0.519 |
0.064 |
| 39 |
263594_at |
AT2G01880
|
ATPAP7, PURPLE ACID PHOSPHATASE 7, PAP7, purple acid phosphatase 7 |
0.513 |
-0.048 |
| 40 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.509 |
0.887 |
| 41 |
264404_at |
AT2G25160
|
CYP82F1, cytochrome P450, family 82, subfamily F, polypeptide 1 |
0.504 |
-0.044 |
| 42 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.503 |
0.107 |
| 43 |
247577_at |
AT5G61290
|
[Flavin-binding monooxygenase family protein] |
0.490 |
-0.060 |
| 44 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
0.490 |
-0.142 |
| 45 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.469 |
0.493 |
| 46 |
247175_at |
AT5G65280
|
GCL1, GCR2-like 1 |
0.464 |
-0.047 |
| 47 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.460 |
0.039 |
| 48 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.457 |
-0.050 |
| 49 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.455 |
-0.090 |
| 50 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
0.445 |
-0.089 |
| 51 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
0.443 |
-0.064 |
| 52 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
0.441 |
-0.042 |
| 53 |
262516_at |
AT1G17190
|
ATGSTU26, glutathione S-transferase tau 26, GSTU26, glutathione S-transferase tau 26 |
0.439 |
-0.071 |
| 54 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.438 |
0.058 |
| 55 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.430 |
0.317 |
| 56 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.429 |
0.152 |
| 57 |
259846_at |
AT1G72140
|
[Major facilitator superfamily protein] |
0.426 |
0.009 |
| 58 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.425 |
1.244 |
| 59 |
259576_at |
AT1G35330
|
[RING/U-box superfamily protein] |
0.413 |
-0.021 |
| 60 |
258652_at |
AT3G09910
|
ATRAB18C, ATRABC2B, RAB GTPase homolog C2B, RABC2b, RAB GTPase homolog C2B |
0.409 |
0.015 |
| 61 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.401 |
0.096 |
| 62 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.399 |
0.130 |
| 63 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.399 |
0.624 |
| 64 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
0.397 |
0.008 |
| 65 |
245586_at |
AT4G14980
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.396 |
-0.025 |
| 66 |
265345_at |
AT2G22680
|
WAVH1, WAV3 homolog 1 |
0.393 |
-0.024 |
| 67 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
0.391 |
0.001 |
| 68 |
250474_at |
AT5G10230
|
ANN7, ANNEXIN 7, ANNAT7, annexin 7 |
0.385 |
-0.043 |
| 69 |
259548_at |
AT1G35260
|
MLP165, MLP-like protein 165 |
0.385 |
-0.041 |
| 70 |
249545_at |
AT5G38030
|
[MATE efflux family protein] |
0.382 |
-0.019 |
| 71 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
0.381 |
0.328 |
| 72 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.379 |
-0.055 |
| 73 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.379 |
0.103 |
| 74 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
0.377 |
0.043 |
| 75 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.375 |
0.056 |
| 76 |
246429_at |
AT5G17450
|
HIPP21, heavy metal associated isoprenylated plant protein 21 |
0.372 |
-0.066 |
| 77 |
267123_at |
AT2G23560
|
ATMES7, ARABIDOPSIS THALIANA METHYL ESTERASE 7, MES7, methyl esterase 7 |
0.366 |
-0.176 |
| 78 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.366 |
-0.234 |
| 79 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
0.361 |
0.033 |
| 80 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.358 |
0.055 |
| 81 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.356 |
-0.339 |
| 82 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.352 |
0.288 |
| 83 |
256935_at |
AT3G22570
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.352 |
-0.020 |
| 84 |
247047_at |
AT5G66650
|
unknown |
0.352 |
-0.150 |
| 85 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
0.349 |
0.006 |
| 86 |
247360_at |
AT5G63450
|
CYP94B1, cytochrome P450, family 94, subfamily B, polypeptide 1 |
0.340 |
-0.037 |
| 87 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
0.339 |
-0.025 |
| 88 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.339 |
0.052 |
| 89 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
0.338 |
-0.224 |
| 90 |
253720_at |
AT4G29270
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.337 |
-0.272 |
| 91 |
250960_at |
AT5G02940
|
unknown |
0.336 |
0.038 |
| 92 |
245243_at |
AT1G44414
|
unknown |
0.335 |
-0.017 |
| 93 |
248855_at |
AT5G46590
|
anac096, NAC domain containing protein 96, NAC096, NAC domain containing protein 96 |
0.327 |
-0.042 |
| 94 |
267592_at |
AT2G39710
|
[Eukaryotic aspartyl protease family protein] |
0.326 |
0.009 |
| 95 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.325 |
0.084 |
| 96 |
254201_at |
AT4G24130
|
unknown |
0.320 |
0.022 |
| 97 |
257517_at |
AT3G16330
|
unknown |
0.320 |
0.058 |
| 98 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.317 |
0.134 |
| 99 |
261266_at |
AT1G26770
|
AT-EXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXPA10, expansin A10, ATHEXP ALPHA 1.1, ARABIDOPSIS THALIANA EXPANSIN ALPHA 1.1, EXP10, EXPANSIN 10, EXPA10, expansin A10 |
0.311 |
0.037 |
| 100 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.310 |
0.042 |
| 101 |
266596_at |
AT2G46150
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.308 |
-0.017 |
| 102 |
248205_at |
AT5G54300
|
unknown |
0.307 |
-0.010 |
| 103 |
262653_at |
AT1G14130
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.304 |
0.074 |
| 104 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
0.303 |
-0.003 |
| 105 |
266977_at |
AT2G39420
|
[alpha/beta-Hydrolases superfamily protein] |
0.303 |
0.059 |
| 106 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
-1.124 |
-0.045 |
| 107 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
-1.117 |
0.014 |
| 108 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-1.094 |
0.014 |
| 109 |
249919_at |
AT5G19250
|
[Glycoprotein membrane precursor GPI-anchored] |
-1.017 |
0.008 |
| 110 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
-0.996 |
-0.063 |
| 111 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.914 |
0.010 |
| 112 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
-0.846 |
0.021 |
| 113 |
248509_at |
AT5G50335
|
unknown |
-0.843 |
-0.161 |
| 114 |
246000_at |
AT5G20820
|
SAUR76, SMALL AUXIN UPREGULATED RNA 76 |
-0.807 |
-0.123 |
| 115 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
-0.784 |
0.191 |
| 116 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.720 |
-0.121 |
| 117 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.687 |
0.024 |
| 118 |
263680_at |
AT1G26930
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.671 |
0.062 |
| 119 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
-0.657 |
0.285 |
| 120 |
247878_at |
AT5G57760
|
unknown |
-0.645 |
-0.083 |
| 121 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.636 |
-0.055 |
| 122 |
247337_at |
AT5G63660
|
LCR74, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 74, PDF2.5 |
-0.572 |
-0.009 |
| 123 |
247474_at |
AT5G62280
|
unknown |
-0.563 |
-0.014 |
| 124 |
246935_at |
AT5G25350
|
EBF2, EIN3-binding F box protein 2 |
-0.546 |
-0.069 |
| 125 |
257981_at |
AT3G20770
|
AtEIN3, EIN3, ETHYLENE-INSENSITIVE3 |
-0.546 |
0.081 |
| 126 |
266302_at |
AT2G27050
|
AtEIL1, EIL1, ETHYLENE-INSENSITIVE3-like 1 |
-0.543 |
-0.009 |
| 127 |
264645_at |
AT1G08940
|
[Phosphoglycerate mutase family protein] |
-0.530 |
0.062 |
| 128 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
-0.502 |
0.137 |
| 129 |
258434_at |
AT3G16770
|
ATEBP, ethylene-responsive element binding protein, EBP, ethylene-responsive element binding protein, ERF72, ETHYLENE RESPONSE FACTOR 72, RAP2.3, RELATED TO AP2 3 |
-0.483 |
0.000 |
| 130 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.475 |
-0.066 |
| 131 |
264510_at |
AT1G09530
|
PAP3, PHYTOCHROME-ASSOCIATED PROTEIN 3, PIF3, phytochrome interacting factor 3, POC1, PHOTOCURRENT 1 |
-0.472 |
0.043 |
| 132 |
262105_at |
AT1G02810
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.451 |
-0.032 |
| 133 |
252118_at |
AT3G51400
|
unknown |
-0.449 |
-0.045 |
| 134 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
-0.438 |
0.088 |
| 135 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
-0.419 |
0.283 |
| 136 |
251918_at |
AT3G54040
|
[PAR1 protein] |
-0.415 |
-0.027 |
| 137 |
257331_at |
ATMG01330
|
ORF107H |
-0.393 |
-0.026 |
| 138 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
-0.390 |
0.049 |
| 139 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
-0.386 |
-0.021 |
| 140 |
246922_at |
AT5G25110
|
CIPK25, CBL-interacting protein kinase 25, SnRK3.25, SNF1-RELATED PROTEIN KINASE 3.25 |
-0.364 |
-0.058 |
| 141 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.361 |
-0.019 |
| 142 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
-0.354 |
0.159 |
| 143 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.327 |
0.118 |
| 144 |
249010_at |
AT5G44580
|
unknown |
-0.326 |
-0.048 |
| 145 |
246390_at |
AT1G77330
|
ACO5, ACC oxidase 5 |
-0.325 |
-0.051 |
| 146 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.316 |
-0.003 |
| 147 |
249087_at |
AT5G44210
|
ATERF-9, ERF DOMAIN PROTEIN- 9, ATERF9, ERF DOMAIN PROTEIN 9, ERF9, erf domain protein 9 |
-0.314 |
-0.095 |
| 148 |
250049_at |
AT5G17780
|
[alpha/beta-Hydrolases superfamily protein] |
-0.309 |
0.012 |
| 149 |
262452_at |
AT1G11210
|
unknown |
-0.309 |
-0.060 |
| 150 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.304 |
0.050 |
| 151 |
248028_at |
AT5G55620
|
unknown |
-0.302 |
-0.068 |
| 152 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
-0.302 |
0.189 |
| 153 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
-0.299 |
-0.018 |
| 154 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
-0.298 |
0.125 |
| 155 |
257217_at |
AT3G14940
|
ATPPC3, phosphoenolpyruvate carboxylase 3, PPC3, phosphoenolpyruvate carboxylase 3 |
-0.291 |
0.028 |
| 156 |
263850_at |
AT2G04480
|
unknown |
-0.291 |
0.016 |
| 157 |
245576_at |
AT4G14770
|
ATTCX2, TESMIN/TSO1-LIKE CXC 2, TCX2, TESMIN/TSO1-like CXC 2 |
-0.289 |
-0.008 |
| 158 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
-0.288 |
0.224 |
| 159 |
263319_at |
AT2G47160
|
AtBOR1, BOR1, REQUIRES HIGH BORON 1 |
-0.287 |
0.019 |
| 160 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
-0.284 |
-0.071 |
| 161 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.283 |
-0.125 |
| 162 |
265396_at |
AT2G21040
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.273 |
-0.055 |
| 163 |
249497_at |
AT5G39220
|
[alpha/beta-Hydrolases superfamily protein] |
-0.272 |
-0.076 |
| 164 |
262046_at |
AT1G79960
|
ATOFP14, OFP14, ovate family protein 14 |
-0.271 |
-0.040 |
| 165 |
245098_at |
AT2G40940
|
ERS, ETHYLENE RESPONSE SENSOR, ERS1, ethylene response sensor 1 |
-0.271 |
0.021 |
| 166 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
-0.264 |
-0.041 |
| 167 |
265302_at |
AT2G13970
|
[Mutator-like transposase family, has a 4.1e-39 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.262 |
-0.003 |
| 168 |
253817_at |
AT4G28310
|
unknown |
-0.259 |
-0.128 |
| 169 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.258 |
-0.080 |
| 170 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
-0.258 |
0.030 |
| 171 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
-0.255 |
0.324 |
| 172 |
261778_at |
AT1G76220
|
unknown |
-0.254 |
-0.012 |
| 173 |
260692_at |
AT1G32430
|
[F-box and associated interaction domains-containing protein] |
-0.254 |
-0.016 |
| 174 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
-0.251 |
0.028 |
| 175 |
249879_at |
AT5G23170
|
[Protein kinase superfamily protein] |
-0.249 |
-0.081 |
| 176 |
261305_at |
AT1G48470
|
GLN1;5, glutamine synthetase 1;5 |
-0.246 |
-0.098 |
| 177 |
247381_at |
AT5G63390
|
[O-fucosyltransferase family protein] |
-0.243 |
0.063 |
| 178 |
254971_at |
AT4G10380
|
AtNIP5;1, NIP5;1, NOD26-like intrinsic protein 5;1, NLM6, NOD26-LIKE MIP 6, NLM8, NOD26-LIKE MIP 8 |
-0.243 |
-0.012 |
| 179 |
247288_at |
AT5G64330
|
JK218, NPH3, NON-PHOTOTROPIC HYPOCOTYL 3, RPT3, ROOT PHOTOTROPISM 3 |
-0.243 |
-0.058 |
| 180 |
257243_at |
AT3G24230
|
[Pectate lyase family protein] |
-0.241 |
-0.001 |
| 181 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.238 |
-0.079 |
| 182 |
264721_at |
AT1G23000
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.238 |
-0.010 |
| 183 |
261480_at |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-0.235 |
-0.032 |
| 184 |
253985_at |
AT4G26220
|
CCoAOMT7, caffeoyl coenzyme A ester O-methyltransferase 7 |
-0.235 |
-0.048 |
| 185 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.235 |
-0.139 |
| 186 |
246663_at |
AT5G35300
|
unknown |
-0.234 |
0.016 |
| 187 |
252419_at |
AT3G47510
|
unknown |
-0.233 |
-0.134 |
| 188 |
264377_at |
AT2G25060
|
AtENODL14, ENODL14, early nodulin-like protein 14 |
-0.233 |
-0.101 |
| 189 |
259919_at |
AT1G72560
|
PSD, PAUSED |
-0.232 |
0.044 |
| 190 |
259324_at |
AT3G05310
|
MIRO3, MIRO-related GTP-ase 3 |
-0.231 |
-0.023 |
| 191 |
262436_at |
AT1G47610
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.229 |
-0.047 |
| 192 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
-0.227 |
-0.022 |
| 193 |
262393_at |
AT1G49490
|
[Leucine-rich repeat (LRR) family protein] |
-0.227 |
0.015 |
| 194 |
257361_at |
AT2G07740
|
[similar to nucleic acid binding / zinc ion binding [Arabidopsis thaliana] (TAIR:AT2G01050.1)] |
-0.227 |
0.007 |
| 195 |
257982_at |
AT3G20780
|
ATTOP6B, topoisomerase 6 subunit B, BIN3, BRASSINOSTEROID INSENSITIVE 3, HYP6, ELONGATED HYPOCOTYL 6, RHL3, ROOT HAIRLESS 3, TOP6B, topoisomerase 6 subunit B |
-0.223 |
0.075 |
| 196 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
-0.222 |
-0.045 |
| 197 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
-0.222 |
0.104 |
| 198 |
255443_at |
AT4G02700
|
SULTR3;2, sulfate transporter 3;2 |
-0.221 |
-0.106 |
| 199 |
246458_at |
AT5G16860
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.220 |
-0.024 |
| 200 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
-0.219 |
0.070 |
| 201 |
259797_at |
AT1G64410
|
unknown |
-0.218 |
-0.001 |
| 202 |
256507_at |
AT1G75150
|
unknown |
-0.217 |
-0.011 |
| 203 |
261607_at |
AT1G49660
|
AtCXE5, carboxyesterase 5, CXE5, carboxyesterase 5 |
-0.216 |
-0.026 |
| 204 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.216 |
-0.090 |
| 205 |
246868_at |
AT5G26010
|
[Protein phosphatase 2C family protein] |
-0.213 |
-0.039 |
| 206 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
-0.213 |
0.017 |
| 207 |
252575_at |
AT3G45440
|
LecRK-I.6, L-type lectin receptor kinase I.6 |
-0.212 |
-0.028 |
| 208 |
261502_at |
AT1G14440
|
AtHB31, homeobox protein 31, FTM2, FLORAL TRANSITION AT THE MERISTEM2, HB31, homeobox protein 31, ZHD4, ZINC FINGER HOMEODOMAIN 4 |
-0.211 |
-0.064 |
| 209 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.211 |
-0.080 |