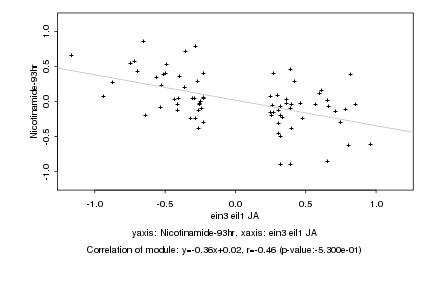
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ein3 eil1 JA |
Log2 signal ratio
Nicotinamide-93hr |
| 1 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
0.956 |
-0.604 |
| 2 |
253060_at |
AT4G37710
|
[VQ motif-containing protein] |
0.849 |
-0.033 |
| 3 |
256382_at |
AT1G66860
|
[Class I glutamine amidotransferase-like superfamily protein] |
0.816 |
0.399 |
| 4 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
0.803 |
-0.620 |
| 5 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
0.782 |
-0.111 |
| 6 |
264636_at |
AT1G65490
|
unknown |
0.742 |
-0.288 |
| 7 |
264635_at |
AT1G65500
|
unknown |
0.710 |
-0.137 |
| 8 |
256603_at |
AT3G28270
|
unknown |
0.658 |
-0.057 |
| 9 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
0.654 |
0.022 |
| 10 |
246001_at |
AT5G20790
|
unknown |
0.650 |
-0.851 |
| 11 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.611 |
0.163 |
| 12 |
249205_at |
AT5G42600
|
MRN1, marneral synthase 1 |
0.596 |
0.115 |
| 13 |
259388_at |
AT1G13420
|
ATST4B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 4B, ST4B, sulfotransferase 4B |
0.564 |
-0.038 |
| 14 |
245226_at |
AT3G29970
|
[B12D protein] |
0.474 |
-0.238 |
| 15 |
259866_at |
AT1G76640
|
CML39, CALMODULIN LIKE 39 |
0.458 |
-0.024 |
| 16 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.414 |
0.295 |
| 17 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
0.395 |
-0.037 |
| 18 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
0.393 |
-0.373 |
| 19 |
260522_x_at |
AT2G41730
|
unknown |
0.391 |
0.459 |
| 20 |
249004_at |
AT5G44570
|
unknown |
0.390 |
-0.889 |
| 21 |
256368_at |
AT1G66800
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.385 |
-0.096 |
| 22 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
0.363 |
0.036 |
| 23 |
266828_at |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
0.361 |
-0.023 |
| 24 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
0.328 |
-0.216 |
| 25 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
0.319 |
-0.058 |
| 26 |
260666_at |
AT1G19300
|
ATGATL1, GATL1, GALACTURONOSYLTRANSFERASE-LIKE 1, GLZ1, GAOLAOZHUANGREN 1, PARVUS, PARVUS |
0.318 |
-0.494 |
| 27 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
0.317 |
-0.893 |
| 28 |
263318_at |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
0.317 |
-0.187 |
| 29 |
262304_at |
AT1G70890
|
MLP43, MLP-like protein 43 |
0.305 |
-0.445 |
| 30 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
0.302 |
-0.301 |
| 31 |
260812_at |
AT1G43650
|
UMAMIT22, Usually multiple acids move in and out Transporters 22 |
0.301 |
-0.127 |
| 32 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.295 |
0.088 |
| 33 |
264940_at |
AT1G60470
|
AtGolS4, galactinol synthase 4, GolS4, galactinol synthase 4 |
0.269 |
-0.147 |
| 34 |
259596_at |
AT1G28130
|
GH3.17 |
0.264 |
0.412 |
| 35 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
0.257 |
-0.043 |
| 36 |
245267_at |
AT4G14060
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.256 |
-0.188 |
| 37 |
251761_at |
AT3G55700
|
[UDP-Glycosyltransferase superfamily protein] |
0.247 |
0.072 |
| 38 |
263594_at |
AT2G01880
|
ATPAP7, PURPLE ACID PHOSPHATASE 7, PAP7, purple acid phosphatase 7 |
0.246 |
-0.156 |
| 39 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
-1.171 |
0.665 |
| 40 |
249364_at |
AT5G40590
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.945 |
0.082 |
| 41 |
263680_at |
AT1G26930
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.875 |
0.272 |
| 42 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.749 |
0.550 |
| 43 |
249454_at |
AT5G39520
|
unknown |
-0.720 |
0.574 |
| 44 |
264645_at |
AT1G08940
|
[Phosphoglycerate mutase family protein] |
-0.703 |
0.431 |
| 45 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
-0.657 |
0.864 |
| 46 |
247337_at |
AT5G63660
|
LCR74, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 74, PDF2.5 |
-0.642 |
-0.195 |
| 47 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.566 |
0.349 |
| 48 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
-0.536 |
-0.077 |
| 49 |
246935_at |
AT5G25350
|
EBF2, EIN3-binding F box protein 2 |
-0.529 |
0.229 |
| 50 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
-0.517 |
0.397 |
| 51 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
-0.501 |
0.411 |
| 52 |
254726_at |
AT4G13660
|
ATPRR2, PRR2, pinoresinol reductase 2 |
-0.495 |
0.536 |
| 53 |
263970_at |
AT2G42850
|
CYP718, cytochrome P450, family 718 |
-0.435 |
0.029 |
| 54 |
246476_at |
AT5G16730
|
[LOCATED IN: chloroplast] |
-0.419 |
-0.121 |
| 55 |
264510_at |
AT1G09530
|
PAP3, PHYTOCHROME-ASSOCIATED PROTEIN 3, PIF3, phytochrome interacting factor 3, POC1, PHOTOCURRENT 1 |
-0.413 |
-0.033 |
| 56 |
246390_at |
AT1G77330
|
ACO5, ACC oxidase 5 |
-0.412 |
0.055 |
| 57 |
258434_at |
AT3G16770
|
ATEBP, ethylene-responsive element binding protein, EBP, ethylene-responsive element binding protein, ERF72, ETHYLENE RESPONSE FACTOR 72, RAP2.3, RELATED TO AP2 3 |
-0.404 |
0.365 |
| 58 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
-0.365 |
0.208 |
| 59 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
-0.361 |
0.716 |
| 60 |
249271_at |
AT5G41790
|
CIP1, COP1-interactive protein 1 |
-0.323 |
-0.234 |
| 61 |
250450_at |
AT5G10280
|
ATMYB64, ATMYB92, myb domain protein 92, MYB92, myb domain protein 92 |
-0.306 |
0.054 |
| 62 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
-0.295 |
0.053 |
| 63 |
258935_at |
AT3G10120
|
unknown |
-0.290 |
-0.230 |
| 64 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
-0.290 |
0.791 |
| 65 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
-0.270 |
0.290 |
| 66 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
-0.267 |
-0.114 |
| 67 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-0.264 |
-0.380 |
| 68 |
257460_at |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
-0.261 |
-0.018 |
| 69 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
-0.256 |
-0.042 |
| 70 |
261117_at |
AT1G75310
|
AUL1, auxilin-like 1 |
-0.255 |
0.009 |
| 71 |
250746_at |
AT5G05880
|
[UDP-Glycosyltransferase superfamily protein] |
-0.248 |
-0.091 |
| 72 |
265913_at |
AT2G25625
|
unknown |
-0.234 |
-0.295 |
| 73 |
253582_at |
AT4G30670
|
[Putative membrane lipoprotein] |
-0.231 |
0.045 |
| 74 |
246493_at |
AT5G16180
|
ATCRS1, ARABIDOPSIS ORTHOLOG OF MAIZE CHLOROPLAST SPLICING FACTOR CRS1, CRS1, ortholog of maize chloroplast splicing factor CRS1 |
-0.228 |
0.070 |
| 75 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.228 |
0.406 |