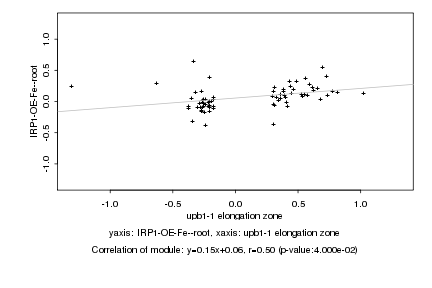
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
upb1-1 elongation zone |
Log2 signal ratio
IRP1-OE-Fe--root |
| 1 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
1.018 |
0.138 |
| 2 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
0.814 |
0.146 |
| 3 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
0.768 |
0.176 |
| 4 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
0.731 |
0.111 |
| 5 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
0.726 |
0.416 |
| 6 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.693 |
0.558 |
| 7 |
265350_at |
AT2G22620
|
[Rhamnogalacturonate lyase family protein] |
0.674 |
0.044 |
| 8 |
259624_at |
AT1G43020
|
unknown |
0.648 |
0.216 |
| 9 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
0.619 |
0.189 |
| 10 |
258143_at |
AT3G18170
|
[Glycosyltransferase family 61 protein] |
0.615 |
0.236 |
| 11 |
252833_at |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
0.589 |
0.282 |
| 12 |
247170_at |
AT5G65530
|
[Protein kinase superfamily protein] |
0.568 |
0.110 |
| 13 |
252537_at |
AT3G45710
|
[Major facilitator superfamily protein] |
0.558 |
0.371 |
| 14 |
265359_at |
AT2G16720
|
ATMYB7, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 7, ATY49, MYB7, myb domain protein 7 |
0.546 |
0.122 |
| 15 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
0.532 |
0.092 |
| 16 |
254015_at |
AT4G26140
|
BGAL12, beta-galactosidase 12 |
0.520 |
0.124 |
| 17 |
254025_at |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.480 |
0.332 |
| 18 |
263437_at |
AT2G28670
|
ESB1, ENHANCED SUBERIN 1 |
0.456 |
0.202 |
| 19 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.446 |
0.136 |
| 20 |
253613_at |
AT4G30320
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.436 |
0.255 |
| 21 |
262743_at |
AT1G29020
|
[Calcium-binding EF-hand family protein] |
0.424 |
0.327 |
| 22 |
258323_at |
AT3G22750
|
[Protein kinase superfamily protein] |
0.410 |
-0.073 |
| 23 |
266968_at |
AT2G39360
|
[Protein kinase superfamily protein] |
0.405 |
-0.013 |
| 24 |
247444_at |
AT5G62630
|
HIPL2, hipl2 protein precursor |
0.399 |
0.070 |
| 25 |
247333_at |
AT5G63600
|
ATFLS5, FLS5, flavonol synthase 5 |
0.384 |
0.105 |
| 26 |
265645_at |
AT2G27370
|
CASP3, Casparian strip membrane domain protein 3 |
0.382 |
0.207 |
| 27 |
257381_at |
AT2G37950
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.380 |
0.176 |
| 28 |
248263_at |
AT5G53370
|
ATPMEPCRF, PECTIN METHYLESTERASE PCR FRAGMENT F, PMEPCRF, pectin methylesterase PCR fragment F |
0.359 |
0.059 |
| 29 |
251918_at |
AT3G54040
|
[PAR1 protein] |
0.354 |
0.124 |
| 30 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
0.336 |
0.020 |
| 31 |
256525_at |
AT1G66180
|
[Eukaryotic aspartyl protease family protein] |
0.324 |
0.079 |
| 32 |
261815_at |
AT1G08320
|
bZIP21, TGA9, TGACG (TGA) motif-binding protein 9 |
0.310 |
-0.063 |
| 33 |
248750_at |
AT5G47530
|
[Auxin-responsive family protein] |
0.306 |
0.228 |
| 34 |
264545_at |
AT1G55670
|
PSAG, photosystem I subunit G |
0.302 |
-0.361 |
| 35 |
246367_at |
AT1G51880
|
RHS6, root hair specific 6 |
0.302 |
-0.034 |
| 36 |
265038_at |
AT1G03920
|
[Protein kinase family protein] |
0.298 |
0.171 |
| 37 |
262049_at |
AT1G80180
|
unknown |
0.289 |
0.084 |
| 38 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
-1.315 |
0.241 |
| 39 |
266276_at |
AT2G29330
|
TRI, tropinone reductase |
-0.633 |
0.304 |
| 40 |
264049_at |
AT2G22390
|
ATRABA4E, RABA4E, RAB GTPase homolog A4E |
-0.380 |
-0.109 |
| 41 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.376 |
-0.072 |
| 42 |
261929_at |
AT1G22460
|
[O-fucosyltransferase family protein] |
-0.354 |
0.051 |
| 43 |
249824_at |
AT5G23380
|
unknown |
-0.347 |
-0.305 |
| 44 |
250455_at |
AT5G09980
|
PROPEP4, elicitor peptide 4 precursor |
-0.337 |
0.644 |
| 45 |
266239_at |
AT2G29530
|
TIM10 |
-0.320 |
0.147 |
| 46 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.308 |
-0.094 |
| 47 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
-0.291 |
-0.026 |
| 48 |
258333_at |
AT3G16000
|
MFP1, MAR binding filament-like protein 1 |
-0.280 |
-0.092 |
| 49 |
248975_at |
AT5G45040
|
CYTC6A, cytochrome c6A |
-0.278 |
-0.152 |
| 50 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
-0.277 |
-0.131 |
| 51 |
257374_at |
AT2G43280
|
[Far-red impaired responsive (FAR1) family protein] |
-0.275 |
0.174 |
| 52 |
249872_at |
AT5G23130
|
[Peptidoglycan-binding LysM domain-containing protein] |
-0.271 |
-0.092 |
| 53 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
-0.267 |
-0.014 |
| 54 |
245947_at |
AT5G19530
|
ACL5, ACAULIS 5 |
-0.267 |
0.021 |
| 55 |
260472_at |
AT1G10990
|
unknown |
-0.261 |
-0.070 |
| 56 |
246396_at |
AT1G58180
|
ATBCA6, A. THALIANA BETA CARBONIC ANHYDRASE 6, BCA6, beta carbonic anhydrase 6 |
-0.260 |
0.042 |
| 57 |
252361_at |
AT3G48490
|
unknown |
-0.258 |
-0.023 |
| 58 |
245123_at |
AT2G47450
|
CAO, CHAOS, CPSRP43, CHLOROPLAST SIGNAL RECOGNITION PARTICLE 43 |
-0.253 |
-0.167 |
| 59 |
249650_at |
AT5G37055
|
ATSWC6, SEF, SERRATED LEAVES AND EARLY FLOWERING |
-0.243 |
0.035 |
| 60 |
261311_at |
AT1G05770
|
[Mannose-binding lectin superfamily protein] |
-0.243 |
-0.033 |
| 61 |
249694_at |
AT5G35790
|
G6PD1, glucose-6-phosphate dehydrogenase 1 |
-0.241 |
-0.370 |
| 62 |
249285_at |
AT5G41960
|
unknown |
-0.217 |
-0.006 |
| 63 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
-0.216 |
-0.078 |
| 64 |
261058_at |
AT1G01370
|
CENH3, CENTROMERIC HISTONE H3, HTR12 |
-0.214 |
-0.059 |
| 65 |
246212_at |
AT4G36930
|
SPT, SPATULA |
-0.214 |
-0.088 |
| 66 |
250718_at |
AT5G06240
|
emb2735, embryo defective 2735 |
-0.212 |
0.016 |
| 67 |
257132_at |
AT3G20230
|
[Ribosomal L18p/L5e family protein] |
-0.209 |
-0.158 |
| 68 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
-0.208 |
0.387 |
| 69 |
246211_at |
AT4G36730
|
AtGBF1, GBF1, G-box binding factor 1 |
-0.192 |
0.015 |
| 70 |
244972_at |
ATCG00680
|
PSBB, photosystem II reaction center protein B |
-0.183 |
0.078 |
| 71 |
260531_at |
AT2G47240
|
CER8, ECERIFERUM 8, LACS1, LONG-CHAIN ACYL-COA SYNTHASE 1 |
-0.182 |
-0.065 |
| 72 |
258969_at |
AT3G10680
|
[HSP20-like chaperones superfamily protein] |
-0.182 |
0.043 |
| 73 |
256805_at |
AT3G20930
|
ORRM1, Organelle RRM protein 1 |
-0.181 |
-0.100 |