|
probeID |
AGICode |
Annotation |
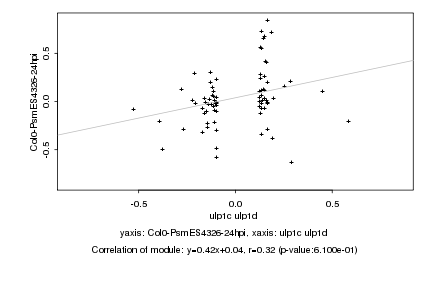
Log2 signal ratio
ulp1c ulp1d |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.582 |
-0.198 |
| 2 |
260145_at |
AT1G52920
|
GCR2, G-PROTEIN COUPLED RECEPTOR 2, GPCR, G protein coupled receptor |
0.446 |
0.111 |
| 3 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
0.288 |
-0.628 |
| 4 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.284 |
0.217 |
| 5 |
264236_at |
AT1G54680
|
unknown |
0.250 |
0.157 |
| 6 |
258781_at |
AT3G11740
|
unknown |
0.195 |
0.033 |
| 7 |
251141_at |
AT5G01075
|
[Glycosyl hydrolase family 35 protein] |
0.191 |
-0.378 |
| 8 |
265327_at |
AT2G18210
|
unknown |
0.186 |
0.724 |
| 9 |
263742_at |
AT2G20625
|
unknown |
0.165 |
-0.001 |
| 10 |
244934_at |
ATCG01080
|
NDHG |
0.164 |
-0.281 |
| 11 |
259618_at |
AT1G48000
|
AtMYB112, myb domain protein 112, MYB112, myb domain protein 112 |
0.161 |
0.851 |
| 12 |
255192_at |
AT4G07380
|
unknown |
0.161 |
-0.017 |
| 13 |
264680_at |
AT1G65510
|
unknown |
0.161 |
0.205 |
| 14 |
265367_at |
AT2G13270
|
unknown |
0.160 |
0.017 |
| 15 |
261785_at |
AT1G08230
|
[Transmembrane amino acid transporter family protein] |
0.158 |
0.409 |
| 16 |
249042_at |
AT5G44350
|
[ethylene-responsive nuclear protein -related] |
0.153 |
0.419 |
| 17 |
266488_at |
AT2G47670
|
PMEI6, PECTIN METHYLESTERASE INHIBITOR 6 |
0.150 |
0.261 |
| 18 |
259324_at |
AT3G05310
|
MIRO3, MIRO-related GTP-ase 3 |
0.149 |
0.116 |
| 19 |
256032_at |
AT1G34090
|
[copia-like retrotransposon family, has a 8.8e-131 P-value blast match to GB:BAA78423 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)GB:BAA78423 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)GB:BAA78423 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)gi|4996361|dbj|BAA78423.1| polyprotein (Arabidopsis thaliana) (Ty1_Copia-element)] |
0.147 |
0.034 |
| 20 |
260151_at |
AT1G52910
|
unknown |
0.146 |
-0.069 |
| 21 |
258880_at |
AT3G06420
|
ATG8H, autophagy 8h |
0.146 |
0.683 |
| 22 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
0.142 |
0.664 |
| 23 |
262339_at |
AT1G64000
|
ATWRKY56, WRKY56, WRKY DNA-binding protein 56 |
0.141 |
0.126 |
| 24 |
248406_at |
AT5G51490
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.137 |
0.013 |
| 25 |
246523_at |
AT5G15850
|
ATCOL1, BBX2, B-box domain protein 2, COL1, CONSTANS-like 1 |
0.134 |
-0.064 |
| 26 |
246841_at |
AT5G26700
|
[RmlC-like cupins superfamily protein] |
0.132 |
0.737 |
| 27 |
249657_at |
AT5G37140
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.131 |
-0.019 |
| 28 |
244996_at |
ATCG00160
|
RPS2, ribosomal protein S2 |
0.130 |
-0.341 |
| 29 |
254537_at |
AT4G19730
|
[Glycosyl hydrolase superfamily protein] |
0.130 |
0.070 |
| 30 |
263744_at |
AT2G21420
|
[IBR domain containing protein] |
0.130 |
0.118 |
| 31 |
249333_at |
AT5G40990
|
GLIP1, GDSL lipase 1 |
0.130 |
0.553 |
| 32 |
260897_at |
AT1G29330
|
AERD2, ARABIDOPSIS ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2, ATERD2, ARABIDOPSIS THALIANA ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2, ERD2, ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2 |
0.128 |
0.566 |
| 33 |
255359_at |
AT4G03950
|
[Nucleotide/sugar transporter family protein] |
0.126 |
0.240 |
| 34 |
260684_at |
AT1G17590
|
NF-YA8, nuclear factor Y, subunit A8 |
0.125 |
0.287 |
| 35 |
252119_at |
AT3G51030
|
ATTRX H1, ARABIDOPSIS THALIANA THIOREDOXIN H-TYPE 1, ATTRX1, thioredoxin H-type 1, TRX1, thioredoxin H-type 1 |
0.125 |
-0.116 |
| 36 |
249958_at |
AT5G18970
|
[AWPM-19-like family protein] |
0.124 |
0.050 |
| 37 |
255892_at |
AT1G17910
|
[Wall-associated kinase family protein] |
0.121 |
0.002 |
| 38 |
262819_at |
AT1G11600
|
CYP77B1, cytochrome P450, family 77, subfamily B, polypeptide 1 |
0.121 |
-0.050 |
| 39 |
246531_at |
AT5G15800
|
AGL2, AGAMOUS-like 2, SEP1, SEPALLATA1 |
0.120 |
0.105 |
| 40 |
263260_at |
AT1G10570
|
OTS2, OVERLY TOLERANT TO SALT 2, ULP1C, UB-LIKE PROTEASE 1C |
-0.528 |
-0.079 |
| 41 |
245025_at |
ATCG00130
|
ATPF |
-0.396 |
-0.200 |
| 42 |
262199_at |
AT1G53800
|
unknown |
-0.381 |
-0.496 |
| 43 |
264268_at |
AT1G60220
|
OTS1, OVERLY TOLERANT TO SALT 1, ULP1D, UB-like protease 1D |
-0.281 |
0.132 |
| 44 |
245227_s_at |
AT1G08410
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.270 |
-0.290 |
| 45 |
244965_at |
ATCG00590
|
ORF31 |
-0.223 |
0.020 |
| 46 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
-0.213 |
0.301 |
| 47 |
265032_at |
AT1G61580
|
ARP2, ARABIDOPSIS RIBOSOMAL PROTEIN 2, RPL3B, R-protein L3 B |
-0.210 |
-0.016 |
| 48 |
266598_at |
AT2G46110
|
KPHMT1, ketopantoate hydroxymethyltransferase 1, PANB1 |
-0.175 |
-0.318 |
| 49 |
245109_at |
AT2G41520
|
DJC62, DNA J protein C62, TPR15, tetratricopeptide repeat 15 |
-0.171 |
-0.071 |
| 50 |
259433_at |
AT1G01570
|
[FUNCTIONS IN: transferase activity, transferring glycosyl groups] |
-0.162 |
0.036 |
| 51 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
-0.161 |
-0.123 |
| 52 |
267511_at |
AT2G45670
|
[calcineurin B subunit-related] |
-0.155 |
0.000 |
| 53 |
266751_at |
AT2G47020
|
[Peptide chain release factor 1] |
-0.151 |
-0.098 |
| 54 |
248679_at |
AT5G48830
|
unknown |
-0.147 |
-0.267 |
| 55 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
-0.145 |
-0.228 |
| 56 |
252087_at |
AT3G52080
|
chx28, cation/hydrogen exchanger 28 |
-0.141 |
-0.024 |
| 57 |
249408_at |
AT5G40330
|
ATMYB23, MYB DOMAIN PROTEIN 23, ATMYBRTF, MYB23, myb domain protein 23 |
-0.137 |
0.029 |
| 58 |
258971_at |
AT3G01990
|
ACR6, ACT domain repeat 6 |
-0.134 |
0.307 |
| 59 |
251262_at |
AT3G62080
|
[SNF7 family protein] |
-0.133 |
0.202 |
| 60 |
251274_at |
AT3G61700
|
unknown |
-0.125 |
-0.029 |
| 61 |
262322_at |
AT1G27590
|
unknown |
-0.122 |
0.069 |
| 62 |
250923_at |
AT5G03455
|
ACR2, ARSENATE REDUCTASE 2, ARATH;CDC25, AtACR2, CDC25 |
-0.121 |
0.153 |
| 63 |
263738_at |
AT1G60060
|
[Serine/threonine-protein kinase WNK (With No Lysine)-related] |
-0.118 |
-0.049 |
| 64 |
256594_at |
AT3G28520
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.118 |
0.105 |
| 65 |
250970_at |
AT5G02770
|
MOS11, modifier of snc1, 11 |
-0.114 |
0.056 |
| 66 |
250674_at |
AT5G07130
|
LAC13, laccase 13 |
-0.112 |
0.014 |
| 67 |
260199_at |
AT1G67590
|
[Remorin family protein] |
-0.111 |
-0.211 |
| 68 |
261710_at |
AT1G32730
|
unknown |
-0.110 |
-0.087 |
| 69 |
259483_at |
AT1G43980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.105 |
-0.012 |
| 70 |
253732_at |
AT4G29140
|
ADS1, ACTIVATED DISEASE SUSCEPTIBILITY 1 |
-0.103 |
-0.032 |
| 71 |
265383_at |
AT2G16780
|
MSI02, MSI2, MULTICOPY SUPPRESSOR OF IRA1 2, NFC02, NFC2, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 2 |
-0.102 |
-0.100 |
| 72 |
252441_at |
AT3G46780
|
PTAC16, plastid transcriptionally active 16 |
-0.102 |
-0.577 |
| 73 |
251083_at |
AT5G01590
|
TIC56, TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 56 |
-0.102 |
-0.479 |
| 74 |
249113_at |
AT5G43790
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.101 |
-0.019 |
| 75 |
251375_at |
AT3G60410
|
unknown |
-0.101 |
0.232 |
| 76 |
258024_at |
AT3G19360
|
[Zinc finger (CCCH-type) family protein] |
-0.101 |
0.051 |
| 77 |
249748_at |
AT5G24620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.100 |
-0.004 |
| 78 |
244966_at |
ATCG00600
|
PETG |
-0.100 |
-0.295 |