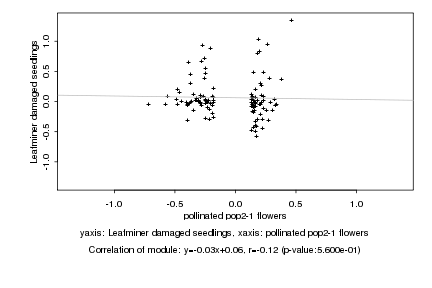
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
pollinated pop2-1 flowers |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
0.455 |
1.360 |
| 2 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.376 |
0.373 |
| 3 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
0.333 |
-0.049 |
| 4 |
257528_at |
AT3G02125
|
unknown |
0.328 |
-0.051 |
| 5 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
0.325 |
-0.053 |
| 6 |
266393_at |
AT2G41260
|
ATM17, M17 |
0.319 |
0.037 |
| 7 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
0.301 |
-0.147 |
| 8 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
0.289 |
-0.016 |
| 9 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.280 |
0.384 |
| 10 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
0.265 |
-0.307 |
| 11 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.261 |
0.951 |
| 12 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
0.256 |
-0.137 |
| 13 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.226 |
0.487 |
| 14 |
253285_at |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
0.226 |
-0.103 |
| 15 |
263837_at |
AT2G04500
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.224 |
0.024 |
| 16 |
263046_at |
AT2G05380
|
GRP3S, glycine-rich protein 3 short isoform |
0.224 |
0.086 |
| 17 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
0.219 |
-0.437 |
| 18 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.217 |
-0.292 |
| 19 |
267303_at |
AT2G30090
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.211 |
-0.016 |
| 20 |
246700_at |
AT5G28030
|
DES1, L-cysteine desulfhydrase 1 |
0.209 |
0.275 |
| 21 |
251598_at |
AT3G57600
|
[Integrase-type DNA-binding superfamily protein] |
0.209 |
0.106 |
| 22 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.206 |
0.315 |
| 23 |
262905_at |
AT1G59730
|
ATH7, thioredoxin H-type 7, TH7, thioredoxin H-type 7 |
0.203 |
-0.044 |
| 24 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
0.202 |
-0.212 |
| 25 |
259054_at |
AT3G03480
|
CHAT, acetyl CoA:(Z)-3-hexen-1-ol acetyltransferase |
0.198 |
0.846 |
| 26 |
254403_at |
AT4G21323
|
[Subtilase family protein] |
0.191 |
-0.047 |
| 27 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
0.185 |
1.035 |
| 28 |
250467_at |
AT5G10100
|
TPPI, trehalose-6-phosphate phosphatase I |
0.176 |
0.032 |
| 29 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
0.175 |
0.802 |
| 30 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
0.175 |
-0.295 |
| 31 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
0.172 |
-0.578 |
| 32 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
0.169 |
-0.411 |
| 33 |
261871_at |
AT1G11440
|
unknown |
0.163 |
0.072 |
| 34 |
253629_at |
AT4G30450
|
[glycine-rich protein] |
0.161 |
0.208 |
| 35 |
265584_at |
AT2G20180
|
PIF1, PHY-INTERACTING FACTOR 1, PIL5, phytochrome interacting factor 3-like 5 |
0.161 |
-0.488 |
| 36 |
257247_at |
AT3G24140
|
FMA, FAMA |
0.161 |
-0.066 |
| 37 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
0.160 |
-0.318 |
| 38 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
0.159 |
-0.385 |
| 39 |
246260_at |
AT1G31820
|
PUT1, POLYAMINE UPTAKE TRANSPORTER 1 |
0.159 |
-0.005 |
| 40 |
251254_at |
AT3G62270
|
BOR2, REQUIRES HIGH BORON 2 |
0.158 |
-0.028 |
| 41 |
262154_at |
AT1G52700
|
[alpha/beta-Hydrolases superfamily protein] |
0.157 |
-0.146 |
| 42 |
248643_at |
AT5G49130
|
[MATE efflux family protein] |
0.152 |
-0.003 |
| 43 |
265248_at |
AT2G43010
|
AtPIF4, PIF4, phytochrome interacting factor 4, SRL2 |
0.147 |
0.025 |
| 44 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
0.147 |
-0.179 |
| 45 |
262039_at |
AT1G80050
|
APT2, adenine phosphoribosyl transferase 2, ATAPT2, PHT1.1 |
0.146 |
-0.419 |
| 46 |
251617_at |
AT3G58000
|
[VQ motif-containing protein] |
0.145 |
-0.085 |
| 47 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
0.144 |
-0.062 |
| 48 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
0.144 |
0.487 |
| 49 |
246106_at |
AT5G28680
|
ANX2, ANXUR2 |
0.140 |
0.045 |
| 50 |
266086_at |
AT2G38060
|
PHT4;2, phosphate transporter 4;2 |
0.137 |
0.080 |
| 51 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
0.137 |
-0.154 |
| 52 |
247038_at |
AT5G67160
|
EPS1, ENHANCED PSEUDOMONAS SUSCEPTIBILTY 1 |
0.136 |
-0.048 |
| 53 |
266644_at |
AT2G29660
|
[zinc finger (C2H2 type) family protein] |
0.135 |
-0.047 |
| 54 |
248864_at |
AT5G46760
|
MYC3 |
0.133 |
0.086 |
| 55 |
247107_at |
AT5G66040
|
STR16, sulfurtransferase protein 16 |
0.130 |
-0.002 |
| 56 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.130 |
-0.070 |
| 57 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
0.130 |
-0.480 |
| 58 |
248540_at |
AT5G50160
|
ATFRO8, FERRIC REDUCTION OXIDASE 8, FRO8, ferric reduction oxidase 8 |
0.130 |
0.042 |
| 59 |
253421_at |
AT4G32340
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.130 |
0.126 |
| 60 |
253724_at |
AT4G29285
|
LCR24, low-molecular-weight cysteine-rich 24 |
-0.720 |
-0.036 |
| 61 |
258590_at |
AT3G04280
|
ARR22, response regulator 22, RR22, response regulator 22 |
-0.578 |
-0.040 |
| 62 |
264401_at |
AT1G61720
|
BAN, BANYULS |
-0.567 |
0.085 |
| 63 |
260066_at |
AT1G73610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.495 |
0.044 |
| 64 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
-0.483 |
0.213 |
| 65 |
245371_at |
AT4G15750
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.482 |
-0.040 |
| 66 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
-0.468 |
0.162 |
| 67 |
264610_at |
AT1G04645
|
[Plant self-incompatibility protein S1 family] |
-0.452 |
0.009 |
| 68 |
266169_at |
AT2G38900
|
[Serine protease inhibitor, potato inhibitor I-type family protein] |
-0.408 |
-0.006 |
| 69 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.398 |
-0.039 |
| 70 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
-0.398 |
-0.312 |
| 71 |
258490_at |
AT3G02670
|
[Glycine-rich protein family] |
-0.396 |
-0.051 |
| 72 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
-0.390 |
0.651 |
| 73 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.387 |
-0.023 |
| 74 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
-0.377 |
0.456 |
| 75 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.377 |
-0.008 |
| 76 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.377 |
0.300 |
| 77 |
262528_at |
AT1G17260
|
AHA10, autoinhibited H(+)-ATPase isoform 10 |
-0.363 |
0.013 |
| 78 |
264184_at |
AT1G54790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.351 |
-0.147 |
| 79 |
245158_at |
AT2G33130
|
RALF18, RALFL18, ralf-like 18 |
-0.350 |
0.118 |
| 80 |
258352_at |
AT3G17600
|
IAA31, indole-3-acetic acid inducible 31 |
-0.336 |
0.029 |
| 81 |
265993_at |
AT2G24160
|
[pseudogene] |
-0.325 |
0.020 |
| 82 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.323 |
0.065 |
| 83 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
-0.310 |
0.027 |
| 84 |
256924_at |
AT3G29590
|
AT5MAT |
-0.310 |
0.013 |
| 85 |
260848_at |
AT1G21850
|
sks8, SKU5 similar 8 |
-0.294 |
-0.027 |
| 86 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.293 |
0.073 |
| 87 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.291 |
-0.016 |
| 88 |
248405_at |
AT5G51480
|
SKS2, SKU5 similar 2 |
-0.290 |
0.113 |
| 89 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
-0.288 |
0.674 |
| 90 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
-0.287 |
-0.052 |
| 91 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
-0.275 |
0.935 |
| 92 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
-0.266 |
0.086 |
| 93 |
262671_at |
AT1G76040
|
CPK29, calcium-dependent protein kinase 29 |
-0.261 |
0.721 |
| 94 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
-0.259 |
0.394 |
| 95 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
-0.252 |
0.559 |
| 96 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.249 |
-0.277 |
| 97 |
254956_at |
AT4G10850
|
AtSWEET7, SWEET7 |
-0.249 |
0.035 |
| 98 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
-0.248 |
0.024 |
| 99 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.248 |
-0.031 |
| 100 |
259385_at |
AT1G13470
|
unknown |
-0.248 |
0.479 |
| 101 |
265954_at |
AT2G37260
|
ATWRKY44, DSL1, DR. STRANGELOVE 1, TTG2, TRANSPARENT TESTA GLABRA 2, WRKY44 |
-0.248 |
-0.032 |
| 102 |
248011_at |
AT5G56300
|
GAMT2, gibberellic acid methyltransferase 2 |
-0.247 |
0.009 |
| 103 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.238 |
-0.093 |
| 104 |
264500_at |
AT1G09370
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.236 |
-0.010 |
| 105 |
257934_at |
AT3G25420
|
scpl21, serine carboxypeptidase-like 21 |
-0.225 |
0.032 |
| 106 |
266321_at |
AT2G46660
|
CYP78A6, cytochrome P450, family 78, subfamily A, polypeptide 6, EOD3, enhancer of da1-1 |
-0.218 |
-0.287 |
| 107 |
246195_at |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
-0.216 |
-0.131 |
| 108 |
258290_at |
AT3G23460
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.210 |
-0.031 |
| 109 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
-0.209 |
0.894 |
| 110 |
260812_at |
AT1G43650
|
UMAMIT22, Usually multiple acids move in and out Transporters 22 |
-0.194 |
-0.183 |
| 111 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
-0.193 |
0.085 |
| 112 |
266736_at |
AT2G46960
|
CYP709B1, cytochrome P450, family 709, subfamily B, polypeptide 1 |
-0.192 |
-0.061 |
| 113 |
249565_at |
AT5G38440
|
[Plant self-incompatibility protein S1 family] |
-0.189 |
-0.004 |
| 114 |
265619_at |
AT2G27320
|
unknown |
-0.188 |
0.021 |
| 115 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.187 |
-0.261 |
| 116 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
-0.186 |
0.073 |
| 117 |
253401_at |
AT4G32870
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.185 |
0.222 |