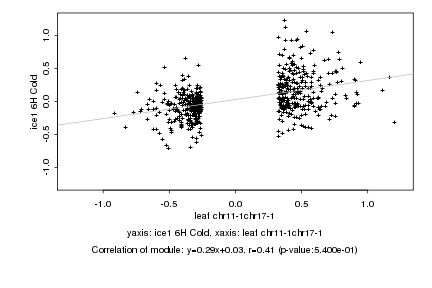
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
leaf chr11-1chr17-1 |
Log2 signal ratio
ice1 6H Cold |
| 1 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
1.201 |
-0.307 |
| 2 |
264872_at |
AT1G24260
|
AGL9, AGAMOUS-like 9, SEP3, SEPALLATA3 |
1.160 |
0.377 |
| 3 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
1.109 |
0.175 |
| 4 |
249454_at |
AT5G39520
|
unknown |
0.945 |
0.600 |
| 5 |
259124_at |
AT3G02310
|
AGL4, AGAMOUS-like 4, SEP2, SEPALLATA 2 |
0.926 |
-0.027 |
| 6 |
252539_at |
AT3G45730
|
unknown |
0.918 |
0.114 |
| 7 |
262719_at |
AT1G43590
|
unknown |
0.910 |
-0.027 |
| 8 |
246889_at |
AT5G25470
|
[AP2/B3-like transcriptional factor family protein] |
0.910 |
0.138 |
| 9 |
256597_at |
AT3G28500
|
[60S acidic ribosomal protein family] |
0.906 |
0.326 |
| 10 |
264638_at |
AT1G65480
|
FT, FLOWERING LOCUS T, RSB8, REDUCED STEM BRANCHING 8 |
0.900 |
-0.073 |
| 11 |
246531_at |
AT5G15800
|
AGL2, AGAMOUS-like 2, SEP1, SEPALLATA1 |
0.894 |
-0.058 |
| 12 |
254574_at |
AT4G19430
|
unknown |
0.893 |
0.342 |
| 13 |
259387_at |
AT1G13370
|
[Histone superfamily protein] |
0.835 |
0.054 |
| 14 |
251304_at |
AT3G61990
|
OMTF3, O-MTase family 3 protein |
0.828 |
0.098 |
| 15 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.803 |
0.516 |
| 16 |
260933_at |
AT1G02470
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.795 |
0.305 |
| 17 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.786 |
0.640 |
| 18 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.778 |
0.758 |
| 19 |
261586_at |
AT1G01640
|
[BTB/POZ domain-containing protein] |
0.767 |
0.289 |
| 20 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.764 |
0.441 |
| 21 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.761 |
0.461 |
| 22 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.756 |
-0.224 |
| 23 |
265471_at |
AT2G37130
|
[Peroxidase superfamily protein] |
0.755 |
0.117 |
| 24 |
264016_at |
AT2G21220
|
SAUR12, SMALL AUXIN UPREGULATED RNA 12 |
0.755 |
-0.028 |
| 25 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.753 |
0.462 |
| 26 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.732 |
0.437 |
| 27 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
0.732 |
1.050 |
| 28 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.731 |
0.050 |
| 29 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.725 |
-0.210 |
| 30 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
0.710 |
-0.270 |
| 31 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
0.707 |
0.264 |
| 32 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.700 |
0.642 |
| 33 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.698 |
0.431 |
| 34 |
251354_at |
AT3G61090
|
[Putative endonuclease or glycosyl hydrolase] |
0.688 |
-0.003 |
| 35 |
247553_at |
AT5G60910
|
AGL8, AGAMOUS-like 8, FUL, FRUITFULL |
0.680 |
-0.061 |
| 36 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.673 |
0.624 |
| 37 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.657 |
0.203 |
| 38 |
256569_at |
AT3G19550
|
unknown |
0.645 |
-0.069 |
| 39 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.645 |
0.159 |
| 40 |
247035_at |
AT5G67110
|
ALC, ALCATRAZ |
0.642 |
-0.119 |
| 41 |
258707_at |
AT3G09480
|
[Histone superfamily protein] |
0.638 |
0.376 |
| 42 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.638 |
0.245 |
| 43 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.633 |
-0.109 |
| 44 |
264823_at |
AT1G03445
|
BSU1, BRI1 SUPPRESSOR 1 |
0.626 |
-0.084 |
| 45 |
255768_at |
AT1G16705
|
[p300/CBP acetyltransferase-related protein-related] |
0.626 |
-0.077 |
| 46 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.626 |
0.063 |
| 47 |
262196_at |
AT1G77870
|
MUB5, membrane-anchored ubiquitin-fold protein 5 precursor |
0.623 |
-0.205 |
| 48 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.622 |
0.323 |
| 49 |
247640_at |
AT5G60610
|
[F-box/RNI-like superfamily protein] |
0.621 |
0.053 |
| 50 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.616 |
0.053 |
| 51 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
0.598 |
-0.146 |
| 52 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.591 |
0.555 |
| 53 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.589 |
0.138 |
| 54 |
253940_at |
AT4G26950
|
unknown |
0.589 |
0.054 |
| 55 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.588 |
0.464 |
| 56 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.585 |
0.784 |
| 57 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.584 |
-0.063 |
| 58 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.583 |
0.429 |
| 59 |
256818_at |
AT3G21420
|
LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1 |
0.573 |
-0.022 |
| 60 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
0.573 |
0.372 |
| 61 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
0.573 |
0.294 |
| 62 |
259293_at |
AT3G11580
|
[AP2/B3-like transcriptional factor family protein] |
0.572 |
-0.013 |
| 63 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
0.572 |
-0.395 |
| 64 |
264580_at |
AT1G05340
|
unknown |
0.570 |
-0.001 |
| 65 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.568 |
0.239 |
| 66 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
0.567 |
0.737 |
| 67 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.564 |
0.199 |
| 68 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.563 |
0.038 |
| 69 |
246025_at |
AT5G21150
|
AGO9, ARGONAUTE 9 |
0.563 |
0.003 |
| 70 |
257824_at |
AT3G25290
|
[Auxin-responsive family protein] |
0.552 |
0.211 |
| 71 |
257460_at |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
0.551 |
0.090 |
| 72 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
0.550 |
-0.393 |
| 73 |
252910_at |
AT4G39590
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.549 |
-0.057 |
| 74 |
260327_at |
AT1G63840
|
[RING/U-box superfamily protein] |
0.548 |
0.091 |
| 75 |
245465_at |
AT4G16590
|
ATCSLA01, cellulose synthase-like A01, ATCSLA1, CELLULOSE SYNTHASE-LIKE A1, CSLA01, CSLA01, cellulose synthase-like A01 |
0.548 |
0.014 |
| 76 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
0.546 |
-0.113 |
| 77 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.544 |
0.384 |
| 78 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.535 |
1.074 |
| 79 |
258362_at |
AT3G14280
|
unknown |
0.531 |
0.436 |
| 80 |
259766_at |
AT1G64360
|
unknown |
0.529 |
-0.197 |
| 81 |
247882_at |
AT5G57785
|
unknown |
0.529 |
-0.190 |
| 82 |
267509_at |
AT2G45660
|
AGL20, AGAMOUS-like 20, ATSOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1, SOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1 |
0.529 |
-0.387 |
| 83 |
246474_at |
AT5G17150
|
[Cystatin/monellin superfamily protein] |
0.524 |
-0.047 |
| 84 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
0.524 |
0.296 |
| 85 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.524 |
0.303 |
| 86 |
265276_at |
AT2G28400
|
unknown |
0.524 |
0.229 |
| 87 |
248668_at |
AT5G48720
|
XRI, X-RAY INDUCED TRANSCRIPT, XRI1, X-RAY INDUCED TRANSCRIPT 1 |
0.521 |
0.078 |
| 88 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
0.519 |
-0.111 |
| 89 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
0.518 |
0.419 |
| 90 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.514 |
0.490 |
| 91 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.513 |
-0.377 |
| 92 |
256874_at |
AT3G26320
|
CYP71B36, cytochrome P450, family 71, subfamily B, polypeptide 36 |
0.512 |
0.458 |
| 93 |
263974_at |
AT2G42720
|
[FBD, F-box, Skp2-like and Leucine Rich Repeat domains containing protein] |
0.512 |
-0.067 |
| 94 |
263164_at |
AT1G03070
|
AtLFG4, LFG4, LIFEGUARD 4 |
0.511 |
-0.066 |
| 95 |
248777_at |
AT5G47920
|
unknown |
0.510 |
0.044 |
| 96 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.509 |
0.539 |
| 97 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.507 |
0.844 |
| 98 |
255430_at |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
0.507 |
0.330 |
| 99 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.507 |
0.365 |
| 100 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.505 |
0.841 |
| 101 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
0.504 |
0.205 |
| 102 |
264832_at |
AT1G03660
|
[Ankyrin-repeat containing protein] |
0.502 |
-0.109 |
| 103 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.499 |
0.667 |
| 104 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.497 |
-0.350 |
| 105 |
260226_at |
AT1G74660
|
MIF1, mini zinc finger 1 |
0.494 |
-0.212 |
| 106 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.494 |
0.303 |
| 107 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
0.491 |
-0.074 |
| 108 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.491 |
0.366 |
| 109 |
251898_at |
AT3G54340
|
AP3, APETALA 3, ATAP3 |
0.488 |
0.163 |
| 110 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.486 |
0.032 |
| 111 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.485 |
0.135 |
| 112 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.485 |
0.822 |
| 113 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.485 |
0.372 |
| 114 |
256442_at |
AT3G10930
|
unknown |
0.483 |
0.441 |
| 115 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.483 |
0.238 |
| 116 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.481 |
-0.255 |
| 117 |
259489_at |
AT1G15790
|
unknown |
0.476 |
0.502 |
| 118 |
257524_at |
AT3G01330
|
DEL3, DP-E2F-like protein 3, E2FF, E2L2, E2F-LIKE 2 |
0.476 |
-0.069 |
| 119 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
0.474 |
0.144 |
| 120 |
252395_at |
AT3G47950
|
AHA4, H(+)-ATPase 4, HA4, H(+)-ATPase 4 |
0.472 |
0.130 |
| 121 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
0.471 |
0.162 |
| 122 |
256789_at |
AT3G13672
|
SINA2 |
0.470 |
0.514 |
| 123 |
245970_at |
AT5G20710
|
BGAL7, beta-galactosidase 7 |
0.469 |
0.211 |
| 124 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
0.468 |
0.160 |
| 125 |
257737_at |
AT3G27440
|
UKL5, uridine kinase-like 5 |
0.466 |
-0.019 |
| 126 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
0.465 |
-0.039 |
| 127 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.465 |
0.149 |
| 128 |
255283_at |
AT4G04620
|
ATG8B, autophagy 8b |
0.465 |
0.278 |
| 129 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.462 |
0.952 |
| 130 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.461 |
0.060 |
| 131 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.458 |
0.278 |
| 132 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.458 |
0.941 |
| 133 |
259142_at |
AT3G10200
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.457 |
-0.228 |
| 134 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
0.457 |
0.430 |
| 135 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.455 |
0.531 |
| 136 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.453 |
-0.081 |
| 137 |
254287_at |
AT4G22960
|
unknown |
0.450 |
-0.044 |
| 138 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
0.449 |
0.108 |
| 139 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
0.449 |
-0.062 |
| 140 |
247869_at |
AT5G57520
|
ATZFP2, ZINC FINGER PROTEIN 2, ZFP2, zinc finger protein 2 |
0.449 |
-0.016 |
| 141 |
247903_at |
AT5G57340
|
unknown |
0.447 |
-0.049 |
| 142 |
246260_at |
AT1G31820
|
PUT1, POLYAMINE UPTAKE TRANSPORTER 1 |
0.445 |
0.297 |
| 143 |
265597_at |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.445 |
0.481 |
| 144 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.444 |
-0.017 |
| 145 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
0.444 |
0.156 |
| 146 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
0.442 |
0.203 |
| 147 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
0.442 |
-0.348 |
| 148 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.442 |
0.567 |
| 149 |
267178_at |
AT2G37750
|
unknown |
0.441 |
0.046 |
| 150 |
261881_at |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
0.439 |
-0.232 |
| 151 |
245401_at |
AT4G17670
|
unknown |
0.439 |
-0.079 |
| 152 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.438 |
0.337 |
| 153 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.436 |
0.707 |
| 154 |
263032_at |
AT1G23850
|
unknown |
0.435 |
0.080 |
| 155 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.434 |
0.422 |
| 156 |
263179_at |
AT1G05710
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.434 |
0.187 |
| 157 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
0.432 |
-0.420 |
| 158 |
262788_at |
AT1G10690
|
unknown |
0.432 |
0.116 |
| 159 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.431 |
0.430 |
| 160 |
267345_at |
AT2G44240
|
unknown |
0.431 |
-0.029 |
| 161 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.431 |
0.552 |
| 162 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.429 |
0.292 |
| 163 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.427 |
0.613 |
| 164 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
0.426 |
0.071 |
| 165 |
251363_at |
AT3G61250
|
AtMYB17, myb domain protein 17, LMI2, LATE MERISTEM IDENTITY2, MYB17, myb domain protein 17 |
0.424 |
-0.129 |
| 166 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.424 |
0.591 |
| 167 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.422 |
0.581 |
| 168 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.420 |
0.928 |
| 169 |
266555_at |
AT2G46270
|
GBF3, G-box binding factor 3 |
0.418 |
0.051 |
| 170 |
256799_at |
AT3G18560
|
unknown |
0.417 |
-0.182 |
| 171 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
0.416 |
0.153 |
| 172 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.411 |
-0.101 |
| 173 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
0.410 |
0.237 |
| 174 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.410 |
0.187 |
| 175 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.409 |
0.482 |
| 176 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
0.409 |
0.191 |
| 177 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.407 |
0.462 |
| 178 |
255726_at |
AT1G25530
|
[Transmembrane amino acid transporter family protein] |
0.405 |
-0.050 |
| 179 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.405 |
-0.180 |
| 180 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.405 |
0.678 |
| 181 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.405 |
0.506 |
| 182 |
251019_at |
AT5G02420
|
unknown |
0.404 |
0.329 |
| 183 |
253776_at |
AT4G28390
|
AAC3, ADP/ATP carrier 3, ATAAC3 |
0.403 |
0.320 |
| 184 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
0.403 |
-0.056 |
| 185 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
0.402 |
0.667 |
| 186 |
248321_at |
AT5G52740
|
[Copper transport protein family] |
0.401 |
0.024 |
| 187 |
251868_at |
AT3G54490
|
RPB5E, RNA polymerase II fifth largest subunit, E |
0.399 |
-0.057 |
| 188 |
260841_at |
AT1G29195
|
unknown |
0.399 |
0.373 |
| 189 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
0.399 |
0.184 |
| 190 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.397 |
-0.437 |
| 191 |
252352_at |
AT3G48185
|
unknown |
0.397 |
-0.018 |
| 192 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
0.394 |
-0.215 |
| 193 |
246888_at |
AT5G26270
|
unknown |
0.394 |
-0.080 |
| 194 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.393 |
0.202 |
| 195 |
253827_at |
AT4G28085
|
unknown |
0.392 |
0.364 |
| 196 |
259846_at |
AT1G72140
|
[Major facilitator superfamily protein] |
0.390 |
-0.052 |
| 197 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
0.390 |
0.057 |
| 198 |
263515_at |
AT2G21640
|
unknown |
0.390 |
0.245 |
| 199 |
249849_at |
AT5G23230
|
NIC2, nicotinamidase 2 |
0.388 |
-0.003 |
| 200 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
0.386 |
-0.089 |
| 201 |
262615_at |
AT1G13950
|
ATELF5A-1, EUKARYOTIC ELONGATION FACTOR 5A-1, EIF-5A, EIF5A, EUKARYOTIC ELONGATION FACTOR 5A, ELF5A-1, eukaryotic elongation factor 5A-1 |
0.386 |
0.298 |
| 202 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.384 |
0.270 |
| 203 |
264761_at |
AT1G61280
|
[Phosphatidylinositol N-acetylglucosaminyltransferase, GPI19/PIG-P subunit] |
0.383 |
-0.055 |
| 204 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
0.381 |
-0.156 |
| 205 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.381 |
0.563 |
| 206 |
262096_at |
AT1G56010
|
anac021, Arabidopsis NAC domain containing protein 21, ANAC022, Arabidopsis NAC domain containing protein 22, NAC1, NAC domain containing protein 1 |
0.380 |
0.287 |
| 207 |
255872_at |
AT2G30360
|
CIPK11, CBL-INTERACTING PROTEIN KINASE 11, PKS5, PROTEIN KINASE SOS2-LIKE 5, SIP4, SOS3-interacting protein 4, SNRK3.22, SNF1-RELATED PROTEIN KINASE 3.22 |
0.380 |
-0.012 |
| 208 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
0.380 |
0.163 |
| 209 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.379 |
0.139 |
| 210 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
0.379 |
0.320 |
| 211 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.379 |
0.303 |
| 212 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
0.377 |
0.126 |
| 213 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.376 |
0.491 |
| 214 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
0.376 |
0.123 |
| 215 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.376 |
-0.067 |
| 216 |
247819_at |
AT5G58350
|
WNK4, with no lysine (K) kinase 4, ZIK2 |
0.375 |
0.242 |
| 217 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.375 |
1.127 |
| 218 |
266548_at |
AT2G35210
|
AGD10, MEE28, MATERNAL EFFECT EMBRYO ARREST 28, RPA, root and pollen arfgap |
0.375 |
-0.053 |
| 219 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
0.375 |
-0.100 |
| 220 |
253689_at |
AT4G29770
|
unknown |
0.374 |
-0.083 |
| 221 |
265902_at |
AT2G25590
|
[Plant Tudor-like protein] |
0.373 |
0.065 |
| 222 |
264635_at |
AT1G65500
|
unknown |
0.371 |
0.936 |
| 223 |
254624_at |
AT4G18580
|
unknown |
0.371 |
0.242 |
| 224 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.370 |
1.245 |
| 225 |
252951_at |
AT4G38700
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.369 |
-0.119 |
| 226 |
258110_at |
AT3G14610
|
CYP72A7, cytochrome P450, family 72, subfamily A, polypeptide 7 |
0.365 |
0.338 |
| 227 |
261890_at |
AT1G80970
|
[XH domain-containing protein] |
0.365 |
0.018 |
| 228 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.364 |
0.791 |
| 229 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.364 |
0.138 |
| 230 |
266977_at |
AT2G39420
|
[alpha/beta-Hydrolases superfamily protein] |
0.363 |
0.059 |
| 231 |
260761_at |
AT1G49150
|
unknown |
0.362 |
0.227 |
| 232 |
259054_at |
AT3G03480
|
CHAT, acetyl CoA:(Z)-3-hexen-1-ol acetyltransferase |
0.362 |
-0.110 |
| 233 |
247345_at |
AT5G63760
|
ARI15, ARIADNE 15, ATARI15, ARABIDOPSIS ARIADNE 15 |
0.362 |
0.240 |
| 234 |
250156_at |
AT5G15170
|
TDP1, tyrosyl-DNA phosphodiesterase 1 |
0.360 |
-0.162 |
| 235 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.359 |
0.258 |
| 236 |
245885_at |
AT5G09440
|
EXL4, EXORDIUM like 4 |
0.358 |
0.059 |
| 237 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.358 |
0.393 |
| 238 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.358 |
0.503 |
| 239 |
245710_at |
AT5G04330
|
CYP84A4, CYTOCHROME P450 84A4 |
0.358 |
-0.058 |
| 240 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
0.357 |
0.217 |
| 241 |
249518_at |
AT5G38610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.357 |
-0.049 |
| 242 |
258021_at |
AT3G19380
|
PUB25, plant U-box 25 |
0.357 |
-0.090 |
| 243 |
251200_at |
AT3G63010
|
ATGID1B, GID1B, GA INSENSITIVE DWARF1B |
0.355 |
0.216 |
| 244 |
252862_at |
AT4G39830
|
[Cupredoxin superfamily protein] |
0.355 |
0.402 |
| 245 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.355 |
0.707 |
| 246 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.354 |
0.051 |
| 247 |
255596_at |
AT4G01720
|
AtWRKY47, WRKY47 |
0.354 |
0.380 |
| 248 |
265012_at |
AT1G24470
|
ATKCR2, KCR2, beta-ketoacyl reductase 2 |
0.354 |
0.058 |
| 249 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.354 |
-0.474 |
| 250 |
257522_at |
AT3G08990
|
[Yippee family putative zinc-binding protein] |
0.353 |
0.268 |
| 251 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
0.352 |
-0.302 |
| 252 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.352 |
0.430 |
| 253 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
0.352 |
-0.103 |
| 254 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.351 |
0.112 |
| 255 |
262646_at |
AT1G62800
|
ASP4, aspartate aminotransferase 4 |
0.350 |
0.008 |
| 256 |
246700_at |
AT5G28030
|
DES1, L-cysteine desulfhydrase 1 |
0.350 |
0.134 |
| 257 |
254266_at |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
0.349 |
0.397 |
| 258 |
251576_at |
AT3G58200
|
[TRAF-like family protein] |
0.349 |
0.264 |
| 259 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.348 |
0.022 |
| 260 |
263914_at |
AT2G36400
|
AtGRF3, growth-regulating factor 3, GRF3, growth-regulating factor 3 |
0.347 |
-0.211 |
| 261 |
258042_at |
AT3G21310
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.346 |
-0.080 |
| 262 |
256063_at |
AT1G07130
|
ATSTN1, STN1 |
0.345 |
-0.011 |
| 263 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.344 |
-0.064 |
| 264 |
251035_at |
AT5G02220
|
SMR4, SIAMESE-RELATED 4 |
0.344 |
0.129 |
| 265 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.344 |
0.138 |
| 266 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.343 |
0.174 |
| 267 |
259555_at |
AT1G21290
|
[copia-like retrotransposon family, has a 3.7e-25 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
0.343 |
-0.092 |
| 268 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
0.342 |
0.378 |
| 269 |
264501_at |
AT1G09390
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.340 |
-0.059 |
| 270 |
246524_at |
AT5G15860
|
ATPCME, prenylcysteine methylesterase, ICME, Isoprenylcysteine methylesterase, PCME, prenylcysteine methylesterase |
0.338 |
0.126 |
| 271 |
264868_at |
AT1G24095
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.336 |
0.240 |
| 272 |
255527_at |
AT4G02360
|
unknown |
0.336 |
0.343 |
| 273 |
267467_at |
AT2G30580
|
BMI1A, DRIP2, DREB2A-interacting protein 2 |
0.336 |
0.232 |
| 274 |
259685_at |
AT1G63090
|
AtPP2-A11, phloem protein 2-A11, PP2-A11, phloem protein 2-A11 |
0.335 |
-0.033 |
| 275 |
251964_at |
AT3G53370
|
[S1FA-like DNA-binding protein] |
0.334 |
0.285 |
| 276 |
258894_at |
AT3G05650
|
AtRLP32, receptor like protein 32, RLP32, receptor like protein 32 |
0.333 |
0.142 |
| 277 |
264752_at |
AT1G23010
|
LPR1, Low Phosphate Root1 |
0.333 |
0.056 |
| 278 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
0.332 |
0.038 |
| 279 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
0.332 |
0.195 |
| 280 |
248539_at |
AT5G50130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.332 |
-0.272 |
| 281 |
262213_at |
AT1G74870
|
[RING/U-box superfamily protein] |
0.331 |
0.193 |
| 282 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.330 |
0.550 |
| 283 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.330 |
0.383 |
| 284 |
257369_at |
AT2G35550
|
ATBPC7, BASIC PENTACYSTEINE 7, BBR, BPC7, basic pentacysteine 7 |
0.330 |
0.126 |
| 285 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.330 |
0.725 |
| 286 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
0.329 |
0.149 |
| 287 |
260943_at |
AT1G45145
|
ATH5, THIOREDOXIN H-TYPE 5, ATTRX5, thioredoxin H-type 5, LIV1, LOCUS OF INSENSITIVITY TO VICTORIN 1, TRX-h5, THIOREDOXIN H-TYPE 5, TRX5, thioredoxin H-type 5 |
0.329 |
0.075 |
| 288 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
0.328 |
-0.150 |
| 289 |
254258_at |
AT4G23410
|
TET5, tetraspanin5 |
0.328 |
-0.003 |
| 290 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
0.325 |
-0.454 |
| 291 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
0.325 |
0.441 |
| 292 |
266064_at |
AT2G18780
|
[F-box and associated interaction domains-containing protein] |
0.325 |
0.128 |
| 293 |
262516_at |
AT1G17190
|
ATGSTU26, glutathione S-transferase tau 26, GSTU26, glutathione S-transferase tau 26 |
0.325 |
0.054 |
| 294 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
0.325 |
0.252 |
| 295 |
258380_at |
AT3G16650
|
PRL2, Pleiotropic regulatory locus 2 |
0.324 |
0.266 |
| 296 |
262601_at |
AT1G15310
|
ATHSRP54A, signal recognition particle 54 kDa subunit, SRP54-1, SIGNAL RECOGNITION PARTICLE 54 KDA SUBUNIT |
0.323 |
0.218 |
| 297 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
0.323 |
-0.525 |
| 298 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.323 |
0.262 |
| 299 |
260237_at |
AT1G74430
|
ATMYB95, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 95, ATMYBCP66, ARABIDOPSIS THALIANA MYB DOMAIN CONTAINING PROTEIN 66, MYB95, myb domain protein 95 |
0.322 |
-0.146 |
| 300 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.322 |
0.981 |
| 301 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.918 |
-0.174 |
| 302 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
-0.838 |
-0.388 |
| 303 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-0.775 |
-0.165 |
| 304 |
264987_at |
AT1G27030
|
unknown |
-0.748 |
0.148 |
| 305 |
254068_at |
AT4G25450
|
ABCB28, ATP-binding cassette B28, ATNAP8, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 8, NAP8, non-intrinsic ABC protein 8 |
-0.725 |
-0.141 |
| 306 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.715 |
-0.107 |
| 307 |
267459_at |
AT2G33850
|
unknown |
-0.669 |
-0.263 |
| 308 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.667 |
-0.037 |
| 309 |
248537_at |
AT5G50100
|
[Putative thiol-disulphide oxidoreductase DCC] |
-0.661 |
-0.106 |
| 310 |
257646_at |
AT3G25740
|
MAP1B, methionine aminopeptidase 1C, MAP1C, METHIONINE AMINOPEPTIDASE 1C |
-0.655 |
0.031 |
| 311 |
246330_at |
AT3G43600
|
AAO2, aldehyde oxidase 2, AO3, aldehyde oxidase 3, AOgamma, Aldehyde oxidase gamma, atAO-2, Arabidopsis thaliana aldehyde oxidase 2, AtAO3, Arabidopsis thaliana aldehyde oxidase 3 |
-0.627 |
0.012 |
| 312 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.626 |
-0.120 |
| 313 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
-0.622 |
-0.108 |
| 314 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.621 |
-0.413 |
| 315 |
263053_at |
AT2G13440
|
[glucose-inhibited division family A protein] |
-0.604 |
-0.200 |
| 316 |
266139_at |
AT2G28085
|
SAUR42, SMALL AUXIN UPREGULATED RNA 42 |
-0.602 |
-0.420 |
| 317 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
-0.600 |
-0.093 |
| 318 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
-0.600 |
0.279 |
| 319 |
246476_at |
AT5G16730
|
[LOCATED IN: chloroplast] |
-0.598 |
0.176 |
| 320 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.579 |
-0.477 |
| 321 |
250233_at |
AT5G13460
|
IQD11, IQ-domain 11 |
-0.576 |
-0.156 |
| 322 |
251646_at |
AT3G57780
|
unknown |
-0.567 |
0.248 |
| 323 |
257057_at |
AT3G15310
|
unknown |
-0.554 |
-0.569 |
| 324 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
-0.554 |
-0.006 |
| 325 |
255243_at |
AT4G05590
|
unknown |
-0.543 |
0.131 |
| 326 |
249271_at |
AT5G41790
|
CIP1, COP1-interactive protein 1 |
-0.539 |
0.045 |
| 327 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
-0.537 |
0.517 |
| 328 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.526 |
-0.185 |
| 329 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.523 |
-0.664 |
| 330 |
258904_at |
AT3G06400
|
CHR11, chromatin-remodeling protein 11 |
-0.517 |
-0.022 |
| 331 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.514 |
-0.038 |
| 332 |
256747_at |
AT3G29180
|
unknown |
-0.510 |
0.002 |
| 333 |
252765_at |
AT3G42800
|
unknown |
-0.508 |
-0.314 |
| 334 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.507 |
-0.701 |
| 335 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.506 |
-0.259 |
| 336 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.506 |
-0.049 |
| 337 |
249046_at |
AT5G44400
|
[FAD-binding Berberine family protein] |
-0.504 |
-0.125 |
| 338 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.497 |
-0.409 |
| 339 |
252391_at |
AT3G47860
|
CHL, chloroplastic lipocalin |
-0.496 |
-0.039 |
| 340 |
257516_at |
AT1G69040
|
ACR4, ACT domain repeat 4 |
-0.491 |
-0.429 |
| 341 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
-0.489 |
-0.314 |
| 342 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
-0.488 |
-0.016 |
| 343 |
267376_at |
AT2G26330
|
ER, ERECTA, QRP1, QUANTITATIVE RESISTANCE TO PLECTOSPHAERELLA 1 |
-0.486 |
-0.460 |
| 344 |
259794_at |
AT1G64330
|
[myosin heavy chain-related] |
-0.475 |
-0.113 |
| 345 |
264698_at |
AT1G70200
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.471 |
-0.096 |
| 346 |
257988_at |
AT3G20870
|
ZTP29, zinc transporter 29 |
-0.465 |
-0.045 |
| 347 |
245859_at |
AT5G28290
|
ATNEK3, NIMA-related kinase 3, NEK3, NIMA-related kinase 3 |
-0.464 |
-0.290 |
| 348 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
-0.460 |
-0.263 |
| 349 |
254303_at |
AT4G22830
|
unknown |
-0.455 |
-0.140 |
| 350 |
246714_at |
AT5G28220
|
[Protein prenylyltransferase superfamily protein] |
-0.454 |
0.073 |
| 351 |
262913_at |
AT1G59960
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.454 |
0.258 |
| 352 |
266175_at |
AT2G02450
|
ANAC034, Arabidopsis NAC domain containing protein 34, ANAC035, NAC domain containing protein 35, AtLOV1, LOV1, LONG VEGETATIVE PHASE 1, NAC035, NAC domain containing protein 35 |
-0.453 |
0.065 |
| 353 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
-0.452 |
-0.082 |
| 354 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.452 |
0.191 |
| 355 |
255128_at |
AT4G08310
|
unknown |
-0.451 |
-0.118 |
| 356 |
252023_at |
AT3G52920
|
unknown |
-0.442 |
-0.253 |
| 357 |
254982_at |
AT4G10470
|
unknown |
-0.439 |
-0.106 |
| 358 |
263431_at |
AT2G22170
|
PLAT2, PLAT domain protein 2 |
-0.434 |
0.142 |
| 359 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.432 |
-0.146 |
| 360 |
266015_at |
AT2G24190
|
SDR2, short-chain dehydrogenase/reductase 2 |
-0.427 |
-0.020 |
| 361 |
260688_at |
AT1G17665
|
unknown |
-0.427 |
0.032 |
| 362 |
255576_at |
AT4G01440
|
UMAMIT31, Usually multiple acids move in and out Transporters 31 |
-0.427 |
-0.285 |
| 363 |
247044_at |
AT5G66850
|
MAPKKK5, mitogen-activated protein kinase kinase kinase 5 |
-0.425 |
-0.156 |
| 364 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
-0.424 |
-0.225 |
| 365 |
250395_at |
AT5G10950
|
[Tudor/PWWP/MBT superfamily protein] |
-0.421 |
0.114 |
| 366 |
264908_at |
AT2G17440
|
PIRL5, plant intracellular ras group-related LRR 5 |
-0.420 |
0.111 |
| 367 |
262533_at |
AT1G17090
|
unknown |
-0.418 |
0.053 |
| 368 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
-0.414 |
-0.188 |
| 369 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
-0.413 |
-0.308 |
| 370 |
261774_at |
AT1G76260
|
DWA2, DWD (DDB1-binding WD40 protein) hypersensitive to ABA 2 |
-0.412 |
0.188 |
| 371 |
251586_at |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
-0.409 |
-0.367 |
| 372 |
245743_at |
AT1G51080
|
unknown |
-0.409 |
-0.356 |
| 373 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.409 |
-0.007 |
| 374 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.409 |
-0.385 |
| 375 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.408 |
-0.335 |
| 376 |
248501_at |
AT5G50440
|
ATMEMB12, MEMB12, membrin 12 |
-0.407 |
0.094 |
| 377 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.406 |
0.205 |
| 378 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.403 |
0.398 |
| 379 |
249340_at |
AT5G41140
|
[Myosin heavy chain-related protein] |
-0.402 |
-0.255 |
| 380 |
251800_at |
AT3G55510
|
RBL, REBELOTE |
-0.402 |
0.061 |
| 381 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
-0.401 |
0.324 |
| 382 |
254692_at |
AT4G17860
|
unknown |
-0.401 |
-0.053 |
| 383 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.394 |
0.185 |
| 384 |
260653_at |
AT1G32440
|
PKp3, plastidial pyruvate kinase 3 |
-0.394 |
-0.013 |
| 385 |
255499_at |
AT4G02730
|
AtWDR5b, WDR5b, human WDR5 (WD40 repeat) homolog b |
-0.394 |
0.143 |
| 386 |
254791_at |
AT4G12910
|
scpl20, serine carboxypeptidase-like 20 |
-0.394 |
-0.131 |
| 387 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
-0.391 |
0.020 |
| 388 |
245830_at |
AT1G57790
|
[F-box family protein] |
-0.390 |
0.112 |
| 389 |
250381_at |
AT5G11610
|
[Exostosin family protein] |
-0.387 |
0.169 |
| 390 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
-0.387 |
0.337 |
| 391 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
-0.385 |
0.667 |
| 392 |
254496_at |
AT4G20070
|
AAH, allantoate amidohydrolase, ATAAH, allantoate amidohydrolase |
-0.384 |
-0.126 |
| 393 |
253021_at |
AT4G38050
|
[Xanthine/uracil permease family protein] |
-0.383 |
-0.042 |
| 394 |
248007_at |
AT5G56260
|
[Ribonuclease E inhibitor RraA/Dimethylmenaquinone methyltransferase] |
-0.382 |
0.048 |
| 395 |
245789_at |
AT1G32090
|
[early-responsive to dehydration stress protein (ERD4)] |
-0.381 |
-0.195 |
| 396 |
257743_at |
AT3G27390
|
unknown |
-0.375 |
0.018 |
| 397 |
262891_at |
AT1G79460
|
ATKS, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE, ATKS1, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE 1, GA2, GA REQUIRING 2, KS, KS1, ENT-KAURENE SYNTHASE 1 |
-0.372 |
-0.188 |
| 398 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.371 |
-0.201 |
| 399 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
-0.370 |
0.183 |
| 400 |
260107_at |
AT1G66430
|
[pfkB-like carbohydrate kinase family protein] |
-0.369 |
-0.039 |
| 401 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
-0.368 |
-0.330 |
| 402 |
249551_at |
AT5G38220
|
[alpha/beta-Hydrolases superfamily protein] |
-0.368 |
-0.118 |
| 403 |
256150_at |
AT1G55120
|
AtcwINV3, 6-fructan exohydrolase, ATFRUCT5, beta-fructofuranosidase 5, FRUCT5, beta-fructofuranosidase 5 |
-0.363 |
0.095 |
| 404 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
-0.363 |
-0.172 |
| 405 |
260011_at |
AT1G68110
|
[ENTH/ANTH/VHS superfamily protein] |
-0.363 |
-0.065 |
| 406 |
267518_at |
AT2G30500
|
NET4B, Networked 4B |
-0.363 |
0.024 |
| 407 |
260806_at |
AT1G78260
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.363 |
-0.095 |
| 408 |
250379_at |
AT5G11590
|
TINY2, TINY2 |
-0.362 |
-0.261 |
| 409 |
246098_at |
AT5G20400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.359 |
0.385 |
| 410 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.358 |
-0.423 |
| 411 |
252165_at |
AT3G50550
|
unknown |
-0.358 |
0.067 |
| 412 |
246063_at |
AT5G19340
|
unknown |
-0.358 |
-0.208 |
| 413 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.357 |
-0.287 |
| 414 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
-0.356 |
-0.270 |
| 415 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.355 |
-0.029 |
| 416 |
255181_at |
AT4G08110
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.2e-66 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.354 |
-0.019 |
| 417 |
264998_at |
AT1G67330
|
unknown |
-0.354 |
0.091 |
| 418 |
245417_at |
AT4G17360
|
[Formyl transferase] |
-0.354 |
-0.278 |
| 419 |
266439_s_at |
AT2G43200
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.351 |
0.257 |
| 420 |
263115_at |
AT1G03055
|
AtD27, A. thaliana homolog of rice D27, D27, DWARF27 |
-0.349 |
-0.351 |
| 421 |
260251_at |
AT1G74250
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.348 |
0.105 |
| 422 |
256602_at |
AT3G28310
|
unknown |
-0.347 |
-0.418 |
| 423 |
265405_at |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.346 |
0.013 |
| 424 |
266735_at |
AT2G46930
|
[Pectinacetylesterase family protein] |
-0.346 |
0.003 |
| 425 |
256149_at |
AT1G55110
|
AtIDD7, indeterminate(ID)-domain 7, IDD7, indeterminate(ID)-domain 7 |
-0.346 |
-0.212 |
| 426 |
254914_at |
AT4G11290
|
[Peroxidase superfamily protein] |
-0.345 |
-0.126 |
| 427 |
265718_at |
AT2G03340
|
WRKY3, WRKY DNA-binding protein 3 |
-0.345 |
-0.032 |
| 428 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.343 |
-0.684 |
| 429 |
247340_at |
AT5G63700
|
[zinc ion binding] |
-0.343 |
-0.091 |
| 430 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
-0.342 |
0.079 |
| 431 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
-0.341 |
-0.020 |
| 432 |
248969_at |
AT5G45310
|
unknown |
-0.340 |
-0.287 |
| 433 |
260416_at |
AT1G69670
|
ATCUL3B, ARABIDOPSIS THALIANA CULLIN 3B, CUL3B, cullin 3B |
-0.340 |
0.056 |
| 434 |
259605_at |
AT1G27910
|
ATPUB45, ARABIDOPSIS THALIANA PLANT U-BOX 45, PUB45, plant U-box 45 |
-0.340 |
-0.247 |
| 435 |
263599_at |
AT2G01830
|
AHK4, ARABIDOPSIS HISTIDINE KINASE 4, ATCRE1, CRE1, CYTOKININ RESPONSE 1, WOL, WOODEN LEG, WOL1, WOODEN LEG 1 |
-0.340 |
-0.047 |
| 436 |
247977_at |
AT5G56850
|
unknown |
-0.339 |
-0.329 |
| 437 |
245756_at |
AT1G35190
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.339 |
0.167 |
| 438 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.339 |
-0.109 |
| 439 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.338 |
-0.359 |
| 440 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-0.338 |
-0.088 |
| 441 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.335 |
-0.315 |
| 442 |
248773_at |
AT5G47820
|
FRA1, FRAGILE FIBER 1 |
-0.335 |
-0.143 |
| 443 |
263556_at |
AT2G16365
|
[F-box family protein] |
-0.334 |
-0.093 |
| 444 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
-0.334 |
-0.066 |
| 445 |
250565_at |
AT5G08000
|
E13L3, glucan endo-1,3-beta-glucosidase-like protein 3, PDCB2, PLASMODESMATA CALLOSE-BINDING PROTEIN 2 |
-0.333 |
-0.135 |
| 446 |
246120_at |
AT5G20360
|
Phox3, Phox3 |
-0.333 |
-0.093 |
| 447 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
-0.331 |
0.100 |
| 448 |
256623_at |
AT3G19960
|
ATM1, myosin 1 |
-0.330 |
-0.201 |
| 449 |
249464_at |
AT5G39710
|
EMB2745, EMBRYO DEFECTIVE 2745 |
-0.330 |
0.109 |
| 450 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
-0.329 |
-0.389 |
| 451 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.328 |
-0.532 |
| 452 |
248538_at |
AT5G50110
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.328 |
-0.090 |
| 453 |
261926_at |
AT1G22530
|
PATL2, PATELLIN 2 |
-0.327 |
-0.078 |
| 454 |
248596_at |
AT5G49330
|
ATMYB111, ARABIDOPSIS MYB DOMAIN PROTEIN 111, MYB111, myb domain protein 111, PFG3, PRODUCTION OF FLAVONOL GLYCOSIDES 3 |
-0.327 |
-0.103 |
| 455 |
250825_at |
AT5G05210
|
[Surfeit locus protein 6] |
-0.325 |
0.007 |
| 456 |
245533_at |
AT4G15130
|
ATCCT2, CCT2, phosphorylcholine cytidylyltransferase2 |
-0.325 |
-0.212 |
| 457 |
256114_at |
AT1G16850
|
unknown |
-0.325 |
-0.329 |
| 458 |
257761_at |
AT3G23090
|
WDL3 |
-0.324 |
-0.088 |
| 459 |
246799_at |
AT5G26940
|
DPD1, defective in pollen organelle DNA degradation1 |
-0.324 |
0.020 |
| 460 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
-0.324 |
-0.120 |
| 461 |
264961_at |
AT1G76950
|
PRAF1 |
-0.322 |
-0.034 |
| 462 |
250429_at |
AT5G10470
|
KAC1, kinesin like protein for actin based chloroplast movement 1, KCA1, KINESIN CDKA;1 ASSOCIATED 1 |
-0.321 |
-0.144 |
| 463 |
264790_at |
AT2G17820
|
AHK1, ATHK1, histidine kinase 1, HK1, histidine kinase 1 |
-0.321 |
0.067 |
| 464 |
265042_at |
AT1G04040
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.320 |
0.033 |
| 465 |
257759_at |
AT3G23070
|
ATCFM3A, CFM3A, CRM family member 3A |
-0.319 |
-0.203 |
| 466 |
263126_at |
AT1G78460
|
[SOUL heme-binding family protein] |
-0.319 |
-0.089 |
| 467 |
256761_at |
AT3G25670
|
[Leucine-rich repeat (LRR) family protein] |
-0.318 |
-0.110 |
| 468 |
250559_at |
AT5G08010
|
unknown |
-0.318 |
0.047 |
| 469 |
253322_at |
AT4G33980
|
unknown |
-0.316 |
-0.123 |
| 470 |
258300_at |
AT3G23340
|
ckl10, casein kinase I-like 10 |
-0.315 |
-0.168 |
| 471 |
266802_at |
AT2G22900
|
[Galactosyl transferase GMA12/MNN10 family protein] |
-0.315 |
0.019 |
| 472 |
258184_at |
AT3G21510
|
AHP1, histidine-containing phosphotransmitter 1 |
-0.315 |
-0.015 |
| 473 |
253263_at |
AT4G34000
|
ABF3, abscisic acid responsive elements-binding factor 3, AtABF3, DPBF5, DC3 PROMOTER-BINDING FACTOR 5 |
-0.314 |
-0.035 |
| 474 |
249361_at |
AT5G40540
|
[Protein kinase superfamily protein] |
-0.312 |
-0.053 |
| 475 |
266964_at |
AT2G39480
|
ABCB6, ATP-binding cassette B6, PGP6, P-glycoprotein 6 |
-0.312 |
0.040 |
| 476 |
245416_at |
AT4G17350
|
unknown |
-0.312 |
-0.173 |
| 477 |
246818_at |
AT5G27270
|
EMB976, EMBRYO DEFECTIVE 976 |
-0.312 |
-0.086 |
| 478 |
245657_at |
AT1G56720
|
[Protein kinase superfamily protein] |
-0.310 |
-0.338 |
| 479 |
251174_at |
AT3G63200
|
PLA IIIB, PLP9, PATATIN-like protein 9 |
-0.308 |
-0.272 |
| 480 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
-0.308 |
-0.182 |
| 481 |
262984_at |
AT1G54460
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.307 |
0.017 |
| 482 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.307 |
-0.254 |
| 483 |
260974_at |
AT1G53440
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.306 |
-0.045 |
| 484 |
258333_at |
AT3G16000
|
MFP1, MAR binding filament-like protein 1 |
-0.306 |
-0.219 |
| 485 |
257598_at |
AT3G24800
|
PRT1, proteolysis 1 |
-0.306 |
-0.014 |
| 486 |
253198_at |
AT4G35360
|
[Uncharacterised conserved protein (UCP030210)] |
-0.305 |
0.076 |
| 487 |
267199_at |
AT2G30990
|
unknown |
-0.304 |
-0.002 |
| 488 |
254487_at |
AT4G20780
|
CML42, calmodulin like 42 |
-0.304 |
0.023 |
| 489 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
-0.303 |
-0.335 |
| 490 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
-0.302 |
-0.330 |
| 491 |
258933_at |
AT3G09980
|
unknown |
-0.302 |
-0.140 |
| 492 |
257814_at |
AT3G25110
|
AtFaTA, fatA acyl-ACP thioesterase, FaTA, fatA acyl-ACP thioesterase |
-0.301 |
-0.042 |
| 493 |
254352_at |
AT4G22320
|
unknown |
-0.299 |
0.043 |
| 494 |
250066_at |
AT5G17930
|
[MIF4G domain-containing protein / MA3 domain-containing protein] |
-0.299 |
-0.011 |
| 495 |
254079_at |
AT4G25730
|
[FtsJ-like methyltransferase family protein] |
-0.299 |
0.202 |
| 496 |
264561_at |
AT1G55810
|
UKL3, uridine kinase-like 3 |
-0.299 |
0.070 |
| 497 |
246247_at |
AT4G36640
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.299 |
0.149 |
| 498 |
248709_at |
AT5G48470
|
PRDA1, PEP-Related Development Arrested 1 |
-0.299 |
0.006 |
| 499 |
251244_at |
AT3G62060
|
[Pectinacetylesterase family protein] |
-0.298 |
-0.115 |
| 500 |
257189_at |
AT3G13222
|
GIP1, GBF-interacting protein 1 |
-0.298 |
-0.061 |
| 501 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.298 |
-0.554 |
| 502 |
262398_at |
AT1G49350
|
[pfkB-like carbohydrate kinase family protein] |
-0.298 |
0.132 |
| 503 |
252073_at |
AT3G51750
|
unknown |
-0.298 |
-0.292 |
| 504 |
254638_at |
AT4G18740
|
[Rho termination factor] |
-0.297 |
-0.220 |
| 505 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.297 |
-0.607 |
| 506 |
253876_at |
AT4G27430
|
CIP7, COP1-interacting protein 7 |
-0.296 |
-0.087 |
| 507 |
248378_at |
AT5G51840
|
unknown |
-0.296 |
0.006 |
| 508 |
266089_at |
AT2G38010
|
[Neutral/alkaline non-lysosomal ceramidase] |
-0.296 |
-0.246 |
| 509 |
250750_at |
AT5G05870
|
UGT76C1, UDP-glucosyl transferase 76C1 |
-0.295 |
-0.103 |
| 510 |
262345_at |
AT1G64180
|
[intracellular protein transport protein USO1-related] |
-0.295 |
-0.034 |
| 511 |
262809_at |
AT1G11720
|
ATSS3, starch synthase 3, SS3, starch synthase 3 |
-0.294 |
-0.102 |
| 512 |
250065_at |
AT5G17910
|
unknown |
-0.294 |
0.064 |
| 513 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
-0.294 |
0.074 |
| 514 |
260598_at |
AT1G55930
|
[CBS domain-containing protein / transporter associated domain-containing protein] |
-0.294 |
-0.074 |
| 515 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
-0.293 |
-0.307 |
| 516 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
-0.293 |
0.107 |
| 517 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
-0.293 |
-0.063 |
| 518 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
-0.292 |
-0.077 |
| 519 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
-0.291 |
-0.322 |
| 520 |
263726_at |
AT2G13610
|
ABCG5, ATP-binding cassette G5 |
-0.291 |
0.055 |
| 521 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
-0.291 |
-0.042 |
| 522 |
267141_at |
AT2G38090
|
[Duplicated homeodomain-like superfamily protein] |
-0.290 |
-0.272 |
| 523 |
257426_at |
AT1G54850
|
[HSP20-like chaperones superfamily protein] |
-0.290 |
-0.079 |
| 524 |
250478_at |
AT5G10250
|
DOT3, DEFECTIVELY ORGANIZED TRIBUTARIES 3 |
-0.290 |
-0.149 |
| 525 |
258560_at |
AT3G06020
|
FAF4, FANTASTIC FOUR 4 |
-0.289 |
-0.147 |
| 526 |
257848_at |
AT3G13030
|
[hAT transposon superfamily protein] |
-0.288 |
-0.084 |
| 527 |
262619_at |
AT1G06550
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
-0.288 |
0.039 |
| 528 |
249515_at |
AT5G38530
|
TSBtype2, tryptophan synthase beta type 2 |
-0.287 |
0.207 |
| 529 |
266774_at |
AT2G29050
|
ATRBL1, RHOMBOID-like 1, RBL1, RHOMBOID-like 1 |
-0.287 |
-0.062 |
| 530 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
-0.287 |
0.245 |
| 531 |
259415_at |
AT1G02330
|
unknown |
-0.287 |
0.020 |
| 532 |
246899_at |
AT5G25590
|
[INVOLVED IN: N-terminal protein myristoylation] |
-0.286 |
-0.307 |
| 533 |
249061_at |
AT5G44550
|
[Uncharacterised protein family (UPF0497)] |
-0.286 |
-0.214 |
| 534 |
247671_at |
AT5G60210
|
RIP5, ROP interactive partner 5 |
-0.286 |
-0.227 |
| 535 |
261307_at |
AT1G48520
|
GATB, GLU-ADT subunit B |
-0.285 |
-0.153 |
| 536 |
255444_at |
AT4G02560
|
LD, luminidependens |
-0.285 |
0.020 |
| 537 |
262503_at |
AT1G21670
|
[LOCATED IN: cell wall, plant-type cell wall] |
-0.284 |
0.085 |
| 538 |
245616_at |
AT4G14480
|
[Protein kinase superfamily protein] |
-0.284 |
0.559 |
| 539 |
260007_at |
AT1G67870
|
[glycine-rich protein] |
-0.284 |
-0.251 |
| 540 |
258046_at |
AT3G21220
|
ATMAP2K_ALPHA, ATMEK5, ARABIDOPSIS THALIANA MITOGEN-ACTIVATED PROTEIN KINASE KINASE 5, ATMKK5, ARABIDOPSIS THALIANA MITOGEN-ACTIVATED PROTEIN KINASE KINASE 5, MAP2K_A, MEK5, MAP KINASE KINASE 5, MKK5, MAP kinase kinase 5 |
-0.284 |
0.108 |
| 541 |
249528_at |
AT5G38720
|
unknown |
-0.284 |
0.076 |
| 542 |
256575_at |
AT3G14790
|
ATRHM3, ARABIDOPSIS THALIANA RHAMNOSE BIOSYNTHESIS 3, RHM3, rhamnose biosynthesis 3 |
-0.283 |
0.038 |
| 543 |
261832_at |
AT1G10650
|
[SBP (S-ribonuclease binding protein) family protein] |
-0.282 |
0.102 |
| 544 |
250751_at |
AT5G05890
|
[UDP-Glycosyltransferase superfamily protein] |
-0.282 |
-0.216 |
| 545 |
253294_at |
AT4G33740
|
unknown |
-0.281 |
-0.169 |
| 546 |
263558_at |
AT2G16380
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.281 |
0.040 |
| 547 |
253078_at |
AT4G36180
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.279 |
-0.157 |
| 548 |
250363_at |
AT5G11390
|
WIT1, WPP domain-interacting protein 1 |
-0.279 |
0.028 |
| 549 |
246468_at |
AT5G17050
|
UGT78D2, UDP-glucosyl transferase 78D2 |
-0.279 |
-0.049 |
| 550 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-0.279 |
-0.304 |
| 551 |
254659_at |
AT4G18240
|
ATSS4, ARABIDOPSIS THALIANA STARCH SYNTHASE 4, SS4, starch synthase 4, SSIV, STARCH SYNTHASE 4 |
-0.278 |
-0.094 |
| 552 |
249469_at |
AT5G39320
|
UDG4, UDP-glucose dehydrogenase 4 |
-0.278 |
-0.113 |
| 553 |
262622_at |
AT1G06510
|
unknown |
-0.278 |
-0.160 |
| 554 |
255008_at |
AT4G10060
|
[Beta-glucosidase, GBA2 type family protein] |
-0.277 |
-0.088 |
| 555 |
260944_at |
AT1G45130
|
AtBGAL5, BGAL5, beta-galactosidase 5 |
-0.277 |
-0.220 |
| 556 |
262538_at |
AT1G17140
|
ICR1, interactor of constitutive active rops 1, RIP1, ROP INTERACTIVE PARTNER 1 |
-0.276 |
-0.462 |
| 557 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.276 |
-0.156 |
| 558 |
261616_at |
AT1G33060
|
ANAC014, NAC 014, NAC014, NAC 014 |
-0.276 |
-0.050 |
| 559 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.275 |
0.211 |
| 560 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.275 |
-0.221 |
| 561 |
255878_at |
AT2G40620
|
[Basic-leucine zipper (bZIP) transcription factor family protein] |
-0.275 |
-0.170 |
| 562 |
263711_at |
AT2G20630
|
PIA1, PP2C induced by AVRRPM1 |
-0.274 |
-0.051 |
| 563 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.274 |
-0.149 |
| 564 |
267197_at |
AT2G30960
|
unknown |
-0.274 |
-0.004 |
| 565 |
247427_at |
AT5G62580
|
[ARM repeat superfamily protein] |
-0.273 |
-0.133 |
| 566 |
255978_at |
AT1G34010
|
unknown |
-0.273 |
-0.073 |
| 567 |
262600_at |
AT1G15340
|
MBD10, methyl-CPG-binding domain 10 |
-0.273 |
-0.221 |
| 568 |
266117_at |
AT2G02170
|
[Remorin family protein] |
-0.272 |
-0.042 |
| 569 |
257073_at |
AT3G19650
|
[cyclin-related] |
-0.272 |
0.048 |
| 570 |
262275_at |
AT1G68710
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.271 |
-0.035 |
| 571 |
257381_at |
AT2G37950
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.271 |
-0.139 |
| 572 |
251198_at |
AT3G62970
|
[zinc finger (C3HC4-type RING finger) family protein] |
-0.270 |
0.009 |
| 573 |
262847_at |
AT1G14840
|
ATMAP70-4, microtubule-associated proteins 70-4, MAP70-4, microtubule-associated proteins 70-4 |
-0.270 |
-0.319 |
| 574 |
257854_at |
AT3G12980
|
ATHPCAT4, HISTONE ACETYLTRANSFERASE OF THE CBP FAMILY 5, HAC5, histone acetyltransferase of the CBP family 5 |
-0.269 |
-0.068 |
| 575 |
254661_at |
AT4G18260
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
-0.269 |
0.059 |
| 576 |
263513_at |
AT2G12400
|
unknown |
-0.269 |
-0.147 |
| 577 |
246313_at |
AT1G31920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.269 |
-0.398 |
| 578 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
-0.268 |
-0.024 |
| 579 |
259595_at |
AT1G28050
|
BBX13, B-box domain protein 13 |
-0.268 |
-0.038 |
| 580 |
245256_at |
AT4G15090
|
FAR1, FAR-RED IMPAIRED RESPONSE 1 |
-0.268 |
-0.097 |
| 581 |
257678_at |
AT3G20420
|
ATRTL2, RNASEIII-LIKE 2, RTL2, RNAse THREE-like protein 2 |
-0.268 |
-0.132 |
| 582 |
255664_at |
AT4G00440
|
TRM15, TON1 Recruiting Motif 15 |
-0.268 |
-0.021 |
| 583 |
265044_at |
AT1G03950
|
VPS2.3, vacuolar protein sorting-associated protein 2.3 |
-0.267 |
0.222 |
| 584 |
246555_at |
AT5G15470
|
GAUT14, galacturonosyltransferase 14 |
-0.267 |
0.147 |
| 585 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
-0.267 |
0.101 |
| 586 |
266796_at |
AT2G02880
|
[mucin-related] |
-0.267 |
0.154 |
| 587 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
-0.266 |
0.142 |
| 588 |
267182_at |
AT2G23360
|
unknown |
-0.266 |
-0.010 |
| 589 |
248106_at |
AT5G55100
|
[SWAP (Suppressor-of-White-APricot)/surp domain-containing protein] |
-0.266 |
-0.018 |
| 590 |
248584_at |
AT5G49960
|
unknown |
-0.266 |
-0.057 |
| 591 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-0.265 |
-0.025 |
| 592 |
259903_at |
AT1G74160
|
TRM4, TON1 Recruiting Motif 4 |
-0.265 |
-0.055 |
| 593 |
256872_at |
AT3G26490
|
[Phototropic-responsive NPH3 family protein] |
-0.265 |
-0.135 |
| 594 |
261593_at |
AT1G33170
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.263 |
-0.507 |
| 595 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.263 |
-0.116 |
| 596 |
257193_at |
AT3G13160
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.262 |
0.027 |
| 597 |
263690_at |
AT1G26960
|
AtHB23, homeobox protein 23, HB23, homeobox protein 23 |
-0.262 |
0.063 |
| 598 |
258245_at |
AT3G29075
|
[glycine-rich protein] |
-0.262 |
0.056 |
| 599 |
260472_at |
AT1G10990
|
unknown |
-0.262 |
-0.083 |
| 600 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
-0.261 |
-0.084 |