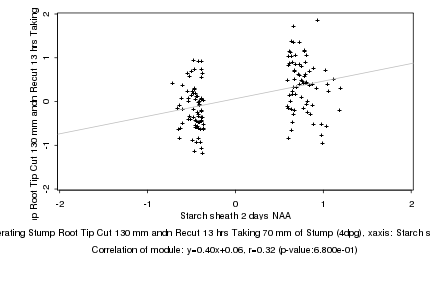
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Starch sheath 2 days NAA |
Log2 signal ratio
Regenerating Stump Root Tip Cut 130 mm andn Recut 13 hrs Taking 70 mm of Stump (4dpg) |
| 1 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
1.188 |
0.306 |
| 2 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
1.179 |
-0.203 |
| 3 |
256890_at |
AT3G23830
|
AtGRP4, GR-RBP4, GRP4, glycine-rich RNA-binding protein 4 |
1.107 |
0.513 |
| 4 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.054 |
0.241 |
| 5 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
1.044 |
0.404 |
| 6 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
1.032 |
-0.565 |
| 7 |
256794_at |
AT3G22230
|
[Ribosomal L27e protein family] |
1.018 |
0.719 |
| 8 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
0.987 |
-0.956 |
| 9 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.975 |
-0.525 |
| 10 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.970 |
-0.775 |
| 11 |
258532_at |
AT3G06700
|
[Ribosomal L29e protein family] |
0.925 |
1.874 |
| 12 |
251486_at |
AT3G59540
|
[Ribosomal L38e protein family] |
0.918 |
0.307 |
| 13 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.884 |
-0.520 |
| 14 |
262880_at |
AT1G64880
|
[Ribosomal protein S5 family protein] |
0.884 |
0.775 |
| 15 |
247607_at |
AT5G60960
|
PNM1, PPR protein localized to the nucleus and mitochondria 1 |
0.873 |
0.394 |
| 16 |
260668_at |
AT1G19530
|
unknown |
0.866 |
-0.079 |
| 17 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.846 |
-0.281 |
| 18 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.836 |
0.379 |
| 19 |
251738_at |
AT3G56150
|
ATEIF3C-1, ATTIF3C1, EIF3C, eukaryotic translation initiation factor 3C, EIF3C-1, TIF3C1 |
0.834 |
0.689 |
| 20 |
251476_at |
AT3G59670
|
unknown |
0.817 |
0.002 |
| 21 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.816 |
-0.244 |
| 22 |
257730_at |
AT3G18420
|
[Protein prenylyltransferase superfamily protein] |
0.807 |
0.439 |
| 23 |
259757_at |
AT1G77510
|
ATPDI6, PROTEIN DISULFIDE ISOMERASE 6, ATPDIL1-2, PDI-like 1-2, PDI6, PROTEIN DISULFIDE ISOMERASE 6, PDIL1-2, PDI-like 1-2 |
0.804 |
0.423 |
| 24 |
261330_at |
AT1G44900
|
ATMCM2, MCM2, MINICHROMOSOME MAINTENANCE 2 |
0.799 |
1.053 |
| 25 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.799 |
-0.051 |
| 26 |
256983_at |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
0.794 |
0.906 |
| 27 |
266822_at |
AT2G44860
|
[Ribosomal protein L24e family protein] |
0.792 |
0.636 |
| 28 |
254726_at |
AT4G13660
|
ATPRR2, PRR2, pinoresinol reductase 2 |
0.791 |
0.424 |
| 29 |
253203_at |
AT4G34710
|
ADC2, arginine decarboxylase 2, ATADC2, SPE2 |
0.779 |
1.170 |
| 30 |
251846_at |
AT3G54560
|
HTA11, histone H2A 11 |
0.777 |
1.166 |
| 31 |
245980_at |
AT5G13140
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.776 |
0.592 |
| 32 |
247032_at |
AT5G67240
|
SDN3, small RNA degrading nuclease 3 |
0.771 |
0.420 |
| 33 |
260235_at |
AT1G74560
|
NRP1, NAP1-related protein 1 |
0.770 |
-0.139 |
| 34 |
254355_at |
AT4G22380
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
0.754 |
0.477 |
| 35 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.750 |
0.802 |
| 36 |
245098_at |
AT2G40940
|
ERS, ETHYLENE RESPONSE SENSOR, ERS1, ethylene response sensor 1 |
0.748 |
0.113 |
| 37 |
249271_at |
AT5G41790
|
CIP1, COP1-interactive protein 1 |
0.742 |
0.495 |
| 38 |
254162_at |
AT4G24440
|
[transcription initiation factor IIA gamma chain / TFIIA-gamma (TFIIA-S)] |
0.727 |
0.410 |
| 39 |
266256_at |
AT2G27710
|
[60S acidic ribosomal protein family] |
0.727 |
1.363 |
| 40 |
247654_at |
AT5G59850
|
[Ribosomal protein S8 family protein] |
0.723 |
0.851 |
| 41 |
245145_at |
AT2G45440
|
DHDPS2, dihydrodipicolinate synthase 2 |
0.722 |
0.603 |
| 42 |
251538_at |
AT3G58660
|
[Ribosomal protein L1p/L10e family] |
0.707 |
0.626 |
| 43 |
264260_at |
AT1G09210
|
AtCRT1b, CRT1b, calreticulin 1b |
0.694 |
0.327 |
| 44 |
252055_at |
AT3G52580
|
[Ribosomal protein S11 family protein] |
0.682 |
1.057 |
| 45 |
256681_at |
AT3G52340
|
ATSPP2, SUCROSE-PHOSPHATASE 2, SPP2, sucrose-6F-phosphate phosphohydrolase 2 |
0.680 |
0.161 |
| 46 |
259403_at |
AT1G17745
|
PGDH, 3-phosphoglycerate dehydrogenase, PGDH2, phosphoglycerate dehydrogenase 2 |
0.677 |
0.850 |
| 47 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.672 |
0.515 |
| 48 |
251029_at |
AT5G02050
|
[Mitochondrial glycoprotein family protein] |
0.670 |
0.696 |
| 49 |
251958_at |
AT3G53560
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.665 |
-0.184 |
| 50 |
253492_at |
AT4G31810
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.662 |
0.714 |
| 51 |
265671_at |
AT2G32060
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
0.658 |
1.732 |
| 52 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
0.657 |
0.343 |
| 53 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.654 |
-0.274 |
| 54 |
246220_at |
AT4G37210
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.650 |
1.364 |
| 55 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
0.649 |
0.246 |
| 56 |
258410_at |
AT3G16780
|
[Ribosomal protein L19e family protein] |
0.649 |
0.904 |
| 57 |
248614_at |
AT5G49560
|
[Putative methyltransferase family protein] |
0.640 |
0.164 |
| 58 |
256442_at |
AT3G10930
|
unknown |
0.639 |
-0.465 |
| 59 |
255657_at |
AT4G00810
|
[60S acidic ribosomal protein family] |
0.636 |
1.389 |
| 60 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.631 |
-0.648 |
| 61 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.631 |
-0.177 |
| 62 |
252625_at |
AT3G44750
|
ATHD2A, HD2A, HISTONE DEACETYLASE 2A, HDA3, histone deacetylase 3, HDT1 |
0.629 |
1.034 |
| 63 |
263047_at |
AT2G17630
|
PSAT2, phosphoserine aminotransferase 2 |
0.622 |
1.126 |
| 64 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.618 |
0.012 |
| 65 |
249466_at |
AT5G39740
|
OLI7, OLIGOCELLULA 7, RPL5B, ribosomal protein L5 B |
0.611 |
0.154 |
| 66 |
248036_at |
AT5G55920
|
OLI2, OLIGOCELLULA 2 |
0.609 |
0.874 |
| 67 |
257177_at |
AT3G23490
|
CYN, cyanase |
0.607 |
1.164 |
| 68 |
253383_at |
AT4G32900
|
[Peptidyl-tRNA hydrolase II (PTH2) family protein] |
0.602 |
-0.140 |
| 69 |
262766_at |
AT1G13160
|
[ARM repeat superfamily protein] |
0.600 |
-0.145 |
| 70 |
261023_at |
AT1G12200
|
FMO, flavin monooxygenase |
0.596 |
-0.845 |
| 71 |
253116_at |
AT4G35850
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.596 |
1.031 |
| 72 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.593 |
0.831 |
| 73 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
0.591 |
0.485 |
| 74 |
248335_at |
AT5G52450
|
[MATE efflux family protein] |
0.591 |
-0.092 |
| 75 |
249227_at |
AT5G42180
|
PER64, peroxidase 64 |
-0.717 |
0.412 |
| 76 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.669 |
-0.144 |
| 77 |
255246_at |
AT4G05640
|
unknown |
-0.651 |
-0.626 |
| 78 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
-0.646 |
-0.823 |
| 79 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.637 |
-0.087 |
| 80 |
262858_at |
AT1G14940
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.634 |
-0.597 |
| 81 |
247663_at |
AT5G60110
|
APUM18, pumilio 18, PUM18, pumilio 18 |
-0.615 |
0.079 |
| 82 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.613 |
-0.180 |
| 83 |
253113_at |
AT4G35985
|
[Senescence/dehydration-associated protein-related] |
-0.607 |
0.379 |
| 84 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
-0.606 |
-0.494 |
| 85 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
-0.556 |
0.249 |
| 86 |
247648_at |
AT5G60020
|
ATLAC17, LAC17, laccase 17 |
-0.554 |
0.658 |
| 87 |
254633_at |
AT4G18640
|
MRH1, morphogenesis of root hair 1 |
-0.543 |
0.021 |
| 88 |
253193_at |
AT4G35380
|
[SEC7-like guanine nucleotide exchange family protein] |
-0.543 |
-0.405 |
| 89 |
260403_at |
AT1G69810
|
ATWRKY36, WRKY36, WRKY DNA-binding protein 36 |
-0.538 |
0.072 |
| 90 |
250233_at |
AT5G13460
|
IQD11, IQ-domain 11 |
-0.529 |
0.576 |
| 91 |
263084_at |
AT2G27180
|
unknown |
-0.529 |
-0.333 |
| 92 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
-0.515 |
-0.392 |
| 93 |
247731_at |
AT5G59590
|
UGT76E2, UDP-glucosyl transferase 76E2 |
-0.511 |
0.157 |
| 94 |
263689_at |
AT1G26820
|
RNS3, ribonuclease 3 |
-0.504 |
0.699 |
| 95 |
249564_at |
AT5G38400
|
unknown |
-0.498 |
-0.884 |
| 96 |
246761_at |
AT5G27980
|
[Seed maturation protein] |
-0.495 |
0.250 |
| 97 |
256860_at |
AT3G23840
|
CER26-LIKE, CER26-LIKE |
-0.488 |
0.955 |
| 98 |
256668_at |
AT3G32190
|
unknown |
-0.486 |
0.200 |
| 99 |
254311_at |
AT4G22440
|
unknown |
-0.484 |
-0.165 |
| 100 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.480 |
-0.360 |
| 101 |
256435_at |
AT3G11180
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.477 |
-1.140 |
| 102 |
250770_at |
AT5G05390
|
LAC12, laccase 12 |
-0.477 |
0.732 |
| 103 |
265754_x_at |
AT2G10640
|
[CACTA-like transposase family (Ptta/En/Spm), has a 7.4e-42 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.474 |
0.302 |
| 104 |
257421_at |
AT1G12030
|
unknown |
-0.471 |
0.288 |
| 105 |
260618_at |
AT1G53230
|
TCP3, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 3 |
-0.463 |
0.069 |
| 106 |
246635_at |
AT1G31720
|
unknown |
-0.461 |
0.190 |
| 107 |
249209_at |
AT5G42620
|
[metalloendopeptidases] |
-0.460 |
-0.534 |
| 108 |
255918_at |
AT5G28570
|
unknown |
-0.457 |
-0.580 |
| 109 |
249852_at |
AT5G23270
|
ATSTP11, SUGAR TRANSPORTER 11, STP11, sugar transporter 11 |
-0.455 |
-0.424 |
| 110 |
257090_at |
AT3G20530
|
[Protein kinase superfamily protein] |
-0.450 |
0.133 |
| 111 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.449 |
-0.927 |
| 112 |
262805_at |
AT1G20900
|
AHL27, AT-hook motif nuclear-localized protein 27, ESC, ESCAROLA, ORE7, ORESARA 7 |
-0.449 |
-0.589 |
| 113 |
251292_at |
AT3G61920
|
unknown |
-0.446 |
-0.455 |
| 114 |
265080_at |
AT1G55570
|
sks12, SKU5 similar 12 |
-0.443 |
-0.570 |
| 115 |
260158_at |
AT1G79910
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.442 |
-0.233 |
| 116 |
246783_at |
AT5G27360
|
SFP2 |
-0.436 |
0.120 |
| 117 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
-0.429 |
-0.596 |
| 118 |
258672_at |
AT3G08570
|
[Phototropic-responsive NPH3 family protein] |
-0.425 |
0.928 |
| 119 |
256662_at |
AT3G11980
|
FAR2, FATTY ACID REDUCTASE 2, MS2, MALE STERILITY 2 |
-0.424 |
-0.022 |
| 120 |
258726_at |
AT3G11745
|
unknown |
-0.423 |
-0.473 |
| 121 |
251252_at |
AT3G62230
|
DAF1, DUO1-activated F-box 1 |
-0.422 |
-0.320 |
| 122 |
265300_at |
AT2G13950
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.422 |
-0.274 |
| 123 |
246826_at |
AT5G26310
|
UGT72E3 |
-0.421 |
-0.827 |
| 124 |
259884_at |
AT1G76390
|
PUB43, plant U-box 43 |
-0.420 |
-0.459 |
| 125 |
259650_at |
AT1G55290
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
-0.418 |
-0.061 |
| 126 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
-0.415 |
-0.083 |
| 127 |
261441_at |
AT1G28470
|
ANAC010, NAC domain containing protein 10, NAC010, NAC domain containing protein 10, SND3, SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 3 |
-0.414 |
0.003 |
| 128 |
254186_at |
AT4G24010
|
ATCSLG1, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G1, CSLG1, cellulose synthase like G1 |
-0.406 |
-0.617 |
| 129 |
264720_at |
AT1G70080
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.401 |
-0.928 |
| 130 |
248355_at |
AT5G52340
|
ATEXO70A2, exocyst subunit exo70 family protein A2, EXO70A2, exocyst subunit exo70 family protein A2 |
-0.397 |
-1.053 |
| 131 |
259961_at |
AT1G53700
|
PK3AT, PROTEIN KINASE 3 ARABIDOPSIS THALIANA, WAG1, WAG 1 |
-0.394 |
0.563 |
| 132 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
-0.392 |
-0.425 |
| 133 |
254263_at |
AT4G23493
|
unknown |
-0.391 |
0.927 |
| 134 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
-0.390 |
0.746 |
| 135 |
258177_at |
AT3G21660
|
[UBX domain-containing protein] |
-0.389 |
-0.435 |
| 136 |
258769_at |
AT3G10870
|
ATMES17, ARABIDOPSIS THALIANA METHYL ESTERASE 17, MES17, methyl esterase 17 |
-0.389 |
-0.359 |
| 137 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.389 |
0.078 |
| 138 |
257752_at |
AT3G18720
|
[F-box family protein] |
-0.389 |
-0.206 |
| 139 |
262031_x_at |
AT1G37160
|
[gypsy-like retrotransposon family (Athila), similar to putative Athila retroelement ORF1 protein GI:4567296 from (Arabidopsis thaliana)] |
-0.388 |
0.048 |
| 140 |
249476_at |
AT5G38910
|
[RmlC-like cupins superfamily protein] |
-0.387 |
-0.202 |
| 141 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.385 |
-0.345 |
| 142 |
247427_at |
AT5G62580
|
[ARM repeat superfamily protein] |
-0.382 |
0.655 |
| 143 |
253587_at |
AT4G30770
|
[Putative membrane lipoprotein] |
-0.377 |
-1.184 |
| 144 |
253501_at |
AT4G32010
|
HSI2-L1, HSL1, HSI2-like 1, VAL2, VP1/ABI3-LIKE 2 |
-0.372 |
0.028 |
| 145 |
265903_at |
AT2G25600
|
AKT6, SPIK, Shaker pollen inward K+ channel |
-0.372 |
-0.619 |
| 146 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
-0.365 |
-0.522 |
| 147 |
247831_at |
AT5G58540
|
[Protein kinase superfamily protein] |
-0.365 |
-0.610 |